Articles
- Page Path
- HOME > Osong Public Health Res Perspect > Volume 4(5); 2013 > Article
-
Original Article
Investigation of Biofilm Formation and its Association with the Molecular and Clinical Characteristics of Methicillin-resistantStaphylococcus aureus - Jeong-Ok Chaa, Jae Il Yooa, Jung Sik Yooa, Hae-Sun Chungb, Sun-Hee Parkc, Hwa Su Kima, Yeong Seon Leea, Gyung Tae Chunga
-
Osong Public Health and Research Perspectives 2013;4(5):225-232.
DOI: https://doi.org/10.1016/j.phrp.2013.09.001
Published online: October 31, 2013
aDivision of Antimicrobial Resistance, Korea National Institute of Health, Osong, Korea
bDepartment of Laboratory Medicine, Ewha Womans University School of Medicine, Seoul, Korea
cDepartment of Internal Medicine, Daejeon St. Mary's Hospital, Daejeon, Korea
- ∗Corresponding author. gtchung@nih.go.kr
© 2013 Published by Elsevier B.V. on behalf of Korea Centers for Disease Control and Prevention.
This is an Open Access article distributed under the terms of the Creative Commons Attribution Non-Commercial License (http://creativecommons.org/licenses/by-nc/3.0) which permits unrestricted non-commercial use, distribution, and reproduction in any medium, provided the original work is properly cited.
Abstract
-
Objectives
- To investigate the biofilm-forming related factors against MRSA bloodstream isolates and evaluates their clinical features and treatment outcomes by biofilm production.
-
Methods
- We collected 126 consecutive methicillin-resistant Staphylococcus aureus (MRSA) causing blood stream infections (BSIs) at 10 tertiary hospitals from 2007 to 2009. We investigated biofilm-forming ability using a microtiter plate assay, and molecular characteristics including multilocus sequence typing, staphylococcal cassette chromosome mec and accessory gene regulator types. We compared the clinical characteristics and outcomes of patients infected with biofilm-forming and non-biofilm-forming MRSA isolates.
-
Results
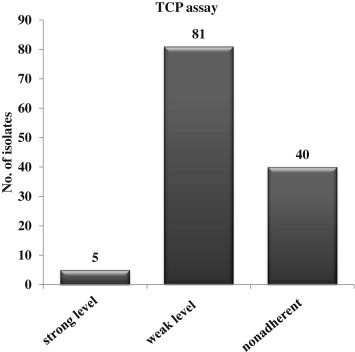
- Of the 126 samples, 86 (68.3%), including 5 strong level (OD570 ≥ 1.0) and 81 weak level (0.2 ≤ OD570 < 1.0), had biofilm-forming capacity. Detection of fibronectinbinding protein in biofilm-forming strains was significantly higher than biofilm non-forming ones (p = 0.001) and three enterotoxin genes (sec-seg-sei) islands had a high frequency regardless of biofilm production. However, biofilm-forming strains were more likely to be multidrug resistant (three or more non-β-lactam antibiotics) than biofilm non-forming ones [79.2% vs. 59.2%, p = 0.015, odds ratio (OR) 2.629, 95% confidence interval (CI) 1.92–5.81]. Clinical features of patients with BSIs caused by biofilm-forming MRSA strains were more likely to be hospital onset [77.9% vs. 60.0%, p = 0.024, OR 2.434, 95% CI 1.11–5.33) and more frequently occurred in patients with use of invasive devices [85.7% vs. 61.2%, p = 0.002, OR 3.879, 95% CI 1.61–8.97]. The other clinical features were compared with the clinical outcomes of the two groups and were not significant (p > 0.05).
-
Conclusion
- Biofilm-forming MRSA strains showed higher frequency of fnbB gene than biofilm non-forming ones and more incidence rates on particular genotypes. And, their patient's features were not significantly different between two groups in this study, except for several clinical factors.
- Biofilms are defined as communities of bacteria encased in a self-synthesized extracellular polymeric matrix that attaches to a biotic or abiotic surface and biofilm-forming staphylococci including Staphylococcus aureus and Staphylococcus epidermidis in gram-positive bacteria have been as one of the major cause of chronic polymer-associated infection [1–3]. Infections involving biofilm-forming bacteria are extremely difficult to eradicate because biofilms impair antibiotic penetration and prevent normal immune responses [4–6]. It has been known that methicillin-resistant S. aureus (MRSA) cause many device-related infections and other chronic infections grow in biofilms or on these devices. Some studies have shown that it is very difficult to treat biofilm-forming staphylococcal infections with antibiotics [7–9]. Moreover, MRSA is the most common cause of nosocomial infections in intensive care units in Korea. Additionally, representative healthcare-associated MRSA strains have progressed to community-associated infections, as has been demonstrated in Korea [10]. However, the prevalence of community-associated MRSA strains in healthcare settings is increasing. Here, we studied the biofilm-forming ability of MRSA blood stream infections (BSIs) and analyzed the relationship between molecular characteristics and their clinical features for MRSA biofilm formers.
Introduction
- 2.1 Bacterial strains and case definition
- We collected MRSA isolates from consecutive unrelated patients in intensive care unit with MRSA BSI at 10 tertiary hospitals from 2007 to 2009. MRSA BSI was considered to be present if one or more blood cultures had positive results, and if clinical signs and course were consistent with MRSA infection. Further case definitions were defined, as previously described [11].
- 2.2 Identification and antimicrobial susceptibility testing
- Identification and antimicrobial susceptibility testing were done using Vitek II (bioMérieux, Craponne, France) or MicroScan Pos Combo Panel Type 6 (Siemens, Munich, Germany) and confirmed by PCR for the presence of mecA gene. We used the following S. aureus strains: ATCC25923, ATCC29213, ATCC43300 (mecA, positive control) from the American Type Culture Collection (Manassas, VA, USA). In addition, antimicrobial susceptibility testing were performed with the disc diffusion method, if needed, according to the Clinical and Laboratory Standards Institute guidelines. S. aureus ATCC 29213 were used as quality control strains for MICs.
- 2.3 Biofilm formation assay
- The biofilm formation assay in microtiter wells was performed as previously described [12]. First, overnight cultures were diluted 1:100 in brain heart infusion broth (Becton Dickinson and Co., Franklin Lakes, NJ, USA) supplemented with 1% glucose. Cell suspensions (200 μL) were transferred to individual wells of a flat-bottom 96-well polystyrene microtiter plate (Nunclon; Nunc, Roskilde, Denmark). After incubation at 35 °C for 48 hours, detached cells were gently rinsed three times with sterile water, and the bacteria that attached to the surface were stained with crystal violet, rewashed, and destained with 1 mL of ethanol-acetone (95:5, vol/vol). A total of 200 μL of the mixed solution was transferred to a 96-well microtiter plate for spectrophotometric analysis at optical density (OD) 570 nm. The absorbance was recorded by Micro-ELISA autoreader (Titertek Multiscan Plus; Labsystems, Helsinki, Finland). Each assay was performed in triplicate and the mean OD570 value of tested wells was applied to biofilm-forming ability. Uninoculated medium was used to determine background. The biofilm formation were divided into three categories in this study, the strains with OD570 < 0.2, 0.2 ≤ OD570 < 1.0, and OD570 ≥ 1.0 were defined as biofilm non-formers, biofilm formers of week level, and strong level, based on the ODs in brain heart infusion broth with and without two supplements. S. aureus SA113 (ATCC35556) and Staphylococcus epidermidis RP62A (ATCC35984), well-characterized biofilm-forming strains, were purchased from American Type Culture Collection for use as positive controls.
- 2.4 Molecular typing
- SCCmec types were determined by using a multiplex PCR strategy according to the method described by Oliveira and de Lancastre [13]. The agr type (1–4) was assigned by PCR as previously described [14]. Multilocus sequence typing was carried out according to protocol previously described [15,16]. Data were analyzed by comparing the database at the multilocus sequence typing website (http://saureus.mlst.net), and the sequence type (ST) for each strain was determined.
- 2.5 Detection of virulence genes
- All 126 strains were analyzed by PCR assay as previously described [12,17]. These included nine staphyloccocal enterotoxin genes (sea, seb, sec, sed, see, seg, seh, sei, sej), three exfoliative toxin genes (eta, etb, etd), six adhesin genes (icaA, icaD, cna, atl, fnbA, fnbB), two surface-associated genes (cap5HK, cap8HK), and two staphylococcal regulators (sarA, arlRS).
- 2.6 Clinical features and outcome
- Medical, laboratory, and pharmacy records were reviewed. Data from patients infected with biofilm-forming and non-forming MRSA isolates were compared. The data collected included age, sex, primary site, and onset of infection. In addition, clinical outcome was assessed for all assessable cases according to biofilm-forming capacity. The outcome measures used were crude mortality, MRSA-related death, and eradication of MRSA. Healthcare-associated risk factors included the following: presence of invasive devices (central venous catheter, urinary catheter, and other indwelling devices), prior use of antibiotics, residence in long-term care facility, prior hospitalization, prior surgery, receipt of hemodialysis, and prior MRSA colonization.
- 2.7 Statistical analysis
- Data analysis was performed using Statistical Package for the Social Sciences (SPSS) Software version 10.0 (SPSS Inc., Chicago, IL, USA) and MedCalc Statistical Software version 10.0.1.0 (MedCalc Software Inc., Mariakerke, Belgium). Statistical significance was assessed via the Pearson χ2 test or the Fisher exact test for categorical variables and the Student t test or the Mann–Whitney U test for continuous variables. Logistic regression analysis was used for multivariate analysis. Variables that achieved a probability of <0.1 in univariate analyses were considered for inclusion in logistic regression models.
Materials and Methods
- 3.1 Antimicrobial susceptibility and biofilm formation
- A total of 126 MRSA isolates from blood cultures were analyzed; 86 strains (68.3%) had biofilm-forming ability with five strong and 81 weak level and the other strains (40/126, 31.7%) did not form biofilms (Figure 1). All isolates were resistant to oxacillin and penicillin. Biofilm-forming isolates were less frequently resistant to oxacillin and penicillin (14.0% vs. 42.5%; p = 0.005), and they were more likely to be multidrug resistant (three or more non-β lactam antibiotics) than biofilm nonforming ones (85.0% vs. 57.5%; p < 0.001; Table 1).
- 3.2 Comparison of genotypic characteristics
- The genetic characteristics of 126 MRSA strains were differed between biofilm-forming and nonforming isolates (Table 2). Most biofilm-forming (55 isolates; 64.0%) and biofilm-nonforming (20 isolates; 50.0%) isolates were of agr group II, with no difference in the distribution of the agr group. The most common SCCmec type was II in both biofilm-forming and nonforming isolates, and the next most frequent type was SCCmec type IV (p = 0.055). Biofilm-forming isolates were more likely to contain ST5 (69.8% vs. 52.5%; p = 0.060), ST239 (8.1% vs. 2.5%; p = 0.434) and significantly less likely to contain ST72 (18.6% vs. 42.5%; p = 0.005) than biofilm nonforming isolates.
- 3.3 Determination of virulence-associated genes
- Table 3 presents the distribution of virulence-associated genes (adhesin-encoding, toxin-encoding, surface-associated, and gene regulators). With a range over 90%, most of the isolates had similar distribution of adhesion genes (icaA, icaD, cna, atl, and fnbA), toxin genes (SEs, hla, and hlb), and staphylococcal regulators (sarA and arlRS) between biofilm-forming and nonforming isolates and both isolates showed low frequency in detection of eta, etb, etd, PVL, and cap8HK. The biofilm-forming isolates were significantly more detected in distribution of fnbB than biofilm-nonforming isolates (74.4% vs. 45.0%; p = 0.001; odds ratio 3.556; 95% confidence interval 1.615–7.827).
- 3.4 Clinical features of patients by biofilm-forming ability
- Epidemiological and clinical data were available for 126 patients and were included in the analysis of factors associated with biofilm formation. Patient characteristics are shown in Table 4. Biofilm-forming isolates were more frequently detected in nosocomial infection (77.9%) than biofilm-nonforming isolates (60%). Among 86 nosocomial infection, biofilm-forming isolates were involved in 67 cases (77.9%), which was higher than community onset cases (22.1%). There were no significant differences in sites of infection. CRI and primary infection were two most common origins of MRSA infection regardless of biofilm-forming ability. As shown in Table 4, there were no significant differences between clinical outcomes of biofilm-forming and nonforming cases for crude mortality and MRSA-related death, in univariate analysis shown in Table 5, significant risk factors associated with healthcare-associated biofilm-forming isolates were the presence of invasive devices (central venous catheter, urinary catheter, and other indwelling devices) and prior hospitalization (p < 0.05). Odds ratios (95% confidence interval) for the variable selected in multivariate analysis were: presence of invasive devices, 3.87 (1.60–8.96); and prior hospitalization, 0.40 (0.18–0.88).
Results
- The ability to form biofilm is a trait that is closely associated with bacterial persistence and virulence, and many persistent and chronic bacterial infections are now believed to be linked to the formation of biofilms [18]. In this study, we analyzed the presence of various virulence genes and adherent proteins against MRSAs causing BSI. The high proportion (about 80–95%) of MRSA isolates in our study could make no significant difference for virulence genes according to biofilm-forming capacity, except for fnbB gene. We found the fnbB gene was to be more frequent among biofilm-producing MRSA strains of ST5 (34/86 isolates) and biofilm non-producing MRSA strains of ST72 (16/40 isolates) respectively (p = 0.005)(data not shown). ST5 strains have been known as frequent genotype in hospital and ST72 in community environment, especially in Asia [11, 19, 20]. In our results, MRSAs of ST5 and ST72 genotypes showed a frequency of fnbB for either biofilm formers or non-formers. This means that the presence of fnbB gene may be correlated with the biofilm-forming ability and MRSA trait from a certain lineage. This presumption could be accordant with a previous study [21], suggesting that FnbB-mediated biofilm development is a common MRSA trait from clonal complex (CC) 8, CC22, and CC45 lineages. However, studies of the relationship of specific STs and biofilm formation require further investigation.
- Epidemiologically biofilm-forming MRSA infection was highly associated with nosocomial infection in this study. Hospital environments may be more suitable for biofilm formation. Various healthcare-associated risk factors are suggested to affect biofilm formation preferable environments to a greater or lesser extent. Our results showed that significant risk factors were the presence of invasive devices and prior hospitalization. Even though not significant statistically, proportions of patients with prior antibiotic use and prior MRSA colonization were higher in biofilm-forming isolates than nonforming isolates. However, among the community onset cases, nearly half of the isolates revealed biofilm-forming activities. This suggests that biofilm formation may be troublesome in community-associated MRSA as well as healthcare-associated MRSA.
- It is widely known that biofilm might play a role in the pathogenesis of device-associated MRSA infections. Particularly, the presence of biofilms on intravascular catheters and their role in catheter-related BSI (CRBSI) is well accepted [22]. We defined CRBSI according to the Infectious Diseases Society of America guidelines [23,24], but the diagnosis of CRBSI remains a major challenge. Although proven CRBSI cases in patients with biofilm-forming isolates were lower than primary BSI in this study, biofilm-formation was significantly associated with the presence of invasive devices, which suggests that invasive devices may be the hidden focus of MRSA BSI. However, this causal relation was not proven and further studies would be necessary to investigate this possibility.
- Biofilm infections are important clinically because bacteria in biofilms exhibit recalcitrance to antimicrobial compounds and persistence in spite of sustained host defenses [25]. Biofilm infection represents a reservoir of dissemination of bacterial infection to other sites in the human body [26]. During infection, attachment is a crucial part of the colonization on host tissues or on indwelling medical devices, whereas detachment is a prerequisite for the dissemination of an infection. Fux et al [20] reported that the detachment of multicellular clumps may explain the high rate of symptomatic metastatic infections seen with S. aureus. They also revealed that nonattached aggregates of bacteria retain the antibiotic resistance seen in biofilms. Collectively, biofilm formation can lead to intractable infection and worse outcome.
- However, significant differences in outcomes between biofilm-forming and nonforming cases were not observed. There are some possible explanations. First, we targeted MRSA isolates that already exhibited a high level of resistance in a significant portion, which may reduce the effects of enhanced antimicrobial resistance due to biofilms. Clinical outcomes are very complex and comprehensive products of various factors including not only bacterial factors but also host factors. Therefore, it is difficult to demonstrate the independent effects of biofilm. In particular, we included patients in intensive care units with MRSA BSI at tertiary hospitals, who generally have very severe status and showed high mortality. By contrast, evaluation of the duration of bacteremia and symptoms would produce different results, as previous studies revealed the association of biofilms and persistent infection [1,25,27], but the information was not submitted.
- In summary, most MRSA isolates related to BSI produced biofilms and their genotypic characteristics have a tendency to having prevalence in some STs and specific genes. Therefore, the patients having biofilm-forming MRSAs seem to be associated with prior use of a medical device and prior hospitalization. These molecular and epidemiological analyses for biofilm-producing MRSAs could be given as basic information for patients who cannot be treated, and may be helpful in determining the possibility of biofilm-related S. aureus infections.
Discussion
-
Acknowledgements
- This study was supported by a grant of the Korea Centers for Disease Control and Prevention (2007-N00294-10).
Acknowledgments
-
This is an Open Access article distributed under the terms of the Creative Commons Attribution Non-Commercial License (http://creativecommons.org/licenses/by-nc/3.0) which permits unrestricted non-commercial use, distribution, and reproduction in any medium, provided the original work is properly cited.
Article information
- 1. Costerton J.W., Stewart P.S., Greenberg E.P.. Bacterial biofilms: a common cause of persistent infections. Science 284(5418). May 21 1999;1318−1322. PMID: 10334980.Article
- 2. Cheung A.L., Eberhardt K., Heinrichs J.H.. Regulation of protein a synthesis by the sar and agr loci of Staphylococcus aureus. Infect Immun 65(6). 1997 Jun;2243−2249. PMID: 9169758.ArticlePubMed
- 3. Patti J.M., Jonsson H., Guss B.. Molecular characterization and expression of a gene encoding a Staphylococcus aureus collagen adhesin. J Biol Chem 267(7). 1992 Mar 5;4766−4772. PMID: 1311320.Article
- 4. Costerton J.W.. Introduction to biofilm. Int J Antimicrob Agents 11(3–4). 1999 May;217−221. [discussion 237–9]. PMID: 10394973.ArticlePubMed
- 5. Stewart P.S., Costerton J.W.. Antibiotic resistance of bacteria in biofilms. Lancet 358(1). 2001 Jul 14;135−138. PMID: 11463434.Article
- 6. Donlan R.M., Costerton J.W.. Biofilms: survival mechanisms of clinically relevant microorganisms. Clin Microbiol Rev 15(2). 2002 Apr;167−193. PMID: 11932229.ArticlePubMed
- 7. Cucarella C., Solano C., Valle J.. Bap, a Staphylococcus aureus surface protein involved in biofilm formation. J Bacteriol 183(9). 2001 May;2888−2896. PMID: 11292810.ArticlePubMed
- 8. O'Neill E., Pozzi C., Houston P.. Association between methicillin susceptibility and biofilm regulation in Staphylococcus aureus isolates from device-related infections. J Clin Microbiol 45(5). 2007 May;1379−1388. PMID: 17329452.ArticlePubMed
- 9. Leid J.G., Shirtliff M.E., Costerton J.W.. Human leukocytes adhere to, penetrate, and respond to Staphylococcus aureus biofilms. Infect Immunity 70(11). 2002 Nov;6339−6345. PMID: 12379713.Article
- 10. Löfdahl S., Guss B., Uhlén M.. Gene for staphylococcal protein A. Proc Natl Acad Sci U S A 80(3). 1983 Feb;697−701. PMID: 6338496.ArticlePubMed
- 11. Park C., Lee D.G., Kim S.W.. Predominance of community-associated methicillin-resistant Staphylococcus aureus strains carrying staphylococcal chromosome cassette mec type IVA in South Korea. J Clin Microbiol 45(12). 2007 Dec;4021−4026. PMID: 17942660.ArticlePubMed
- 12. Ando E., Monden K., Mitsuhata R.. Biofilm formation among methicillin-resistant Staphylococcus aureus isolates from patients with urinary tract infection. Acta Med Okayama 58(4). 2004 Aug;207−214. PMID: 15551758.PubMed
- 13. Oliveira D.C., de Lencastre H.. Multiplex PCR strategy for rapid identification of structural types and variants of the mec element in methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother 46(7). 2002 Jul;2155−2161. PMID: 12069968.ArticlePubMed
- 14. Lina G., Boutite F., Tristan A.. Bacterial competition for human nasal cavity colonization: role of staphylococcal agr alleles. Appl Environ Microbiol 69(1). 2003 Jan;18−23. PMID: 12513972.ArticlePubMed
- 15. Enright M.C., Day N.P., Davies C.E.. Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J Clin Microbiol 38(3). 2000 Mar;1008−1015. PMID: 10698988.ArticlePubMed
- 16. Feil E.J., Li B.C., Aanensen D.M.. eBurst: inferring patterns of evolutionary descent among clusters of related bacterial genotypes from multilocus sequence typing data. J Bacteriol 186(5). 2004 Mar;1518−1530. PMID: 14973027.ArticlePubMed
- 17. Mehrotra M., Wang G., Johnson W.M.. Multiplex PCR for detection of genes for Staphylococcus aureus enterotoxins, exfoliative toxins, toxic shock syndrome toxin 1, and methicillin resistance. J Clin Microbiol 38(3). 2000 Mar;1032−1035. PMID: 10698991.ArticlePubMed
- 18. Mohamed J.A., Huang D.B., Jiang Z.D.. Association of putative enteroaggregative Escherichia coli virulence genes and biofilm production in isolates from travelers to developing countries. J Clin Microbiol 45(1). 2007 Jan;121−126. PMID: 17093030.ArticlePubMed
- 19. Bae I.G., Kim J.S., Kim S.. Genetic correlation of community-associated methicillin-resistant Staphylococcus aureus strains from carriers and from patients with clinical infection in one region of Korea. J Korean Med Sci 25(2). 2010 Feb;197−202. PMID: 20119570.ArticlePubMed
- 20. Fux C.A., Wilson S., Stoodley P.. Detachment characteristics and oxacillin resistance of Staphylococcus aureus biofilm emboli in an in vitro catheter infection model. J Bacteriol 186(14). 2004 Jul;4486−4491. PMID: 15231780.ArticlePubMed
- 21. O'Neill E., Pozzi C., Houston P.. A novel Staphylococcus aureus biofilm phenotype mediated by the fibronectin-binding proteins, fnbpa and fnbpb. J Bacteriol 190(11). 2008 Jun;3835−3850. PMID: 18375547.ArticlePubMed
- 22. Donlan R.M.. Biofilm elimination on intravascular catheters: important considerations for the infectious disease practitioner. Clin Infect Dis 52(8). 2011 Apr 15;1038−1045. PMID: 21460321.Article
- 23. Mermel L.A., Farr B.M., Sherertz R.J.. Guidelines for the management of intravascular catheter-related infections. Clin Infect Dis 32(3). 2001 May–Jun;1249−1272. PMID: 11303260.ArticlePubMed
- 24. Mermel L.A., Allon M., Bouza E.. Clinical practice guidelines for the diagnosis and management of intravascular catheter-related infection: 2009 update by the Infectious Diseases Society of America. Clin Infect Dis 49(1). 2009 Jul;1−45. PMID: 19489710.ArticlePubMed
- 25. Hall-Stoodley L., Stoodley P.. Evolving concepts in biofilm infections. Cell Microbiol 11(7). 2009 Jul;1034−1043. PMID: 19374653.ArticlePubMed
- 26. Otto M.. Staphylococcal biofilms. Curr Top Microbiol Immunol 322:2008;207−228. PMID: 18453278.ArticlePubMedPMC
- 27. Dimitriou G., Fouzas S., Giormezis N.. Clinical and microbiological profile of persistent coagulase-negative staphylococcal bacteraemia in neonates. Clin Microbiol Infect 17(11). 2011 Nov;1684−1690. PMID: 21463392.ArticlePubMed
References
The percentages refer to the percentage of patients within each Staphylococcus aureus subset with the indicated virulence gene.
*The p values comparing the values for the two groups were determined using a two-sided Fisher's exact test (p < 0.05). As determined by the Fisher exact test; all p values shown in this table (referring to a comparison of values for biofilm-forming and non-forming isolates) are statistically significant with a false discovery rate of < 20%.
Figure & Data
References
Citations

- Antibacterial Activity of Boron Compounds Against Biofilm-Forming Pathogens
Ozgur Celebi, Demet Celebi, Sumeyye Baser, Elif Aydın, Erva Rakıcı, Serpil Uğraş, Pınar Ağyar Yoldaş, Nurcan Kılıç Baygutalp, A. M. Abd El-Aty
Biological Trace Element Research.2024; 202(1): 346. CrossRef - Boron Compound–Based Treatments Against Multidrug-Resistant Bacterial Infections in Lung Cancer In Vitro Model
Demet Celebı, Ozgur Celebı, Elif Aydin, Sumeyye Baser, Mustafa Can Güler, Serkan Yildirim, Ali Taghizadehghalehjoughi
Biological Trace Element Research.2024; 202(1): 145. CrossRef - Liposomes-Based Drug Delivery Systems of Anti-Biofilm Agents to Combat Bacterial Biofilm Formation
Zinb Makhlouf, Amaal Abdulraqeb Ali, Mohammad Hussein Al-Sayah
Antibiotics.2023; 12(5): 875. CrossRef - Staphylococcus aureus İzolatlarının Biyofilm Oluşturma Özelliklerinin Karşılaştırılması
Demet GÜR VURAL, İlknur BIYIK, Elif Gülsüm TORUN, Yeliz TANRIVERDİ ÇAYCI, Kemal BİLGİN, Asuman BIRINCI
Sağlık Bilimlerinde Değer.2023; 13(2): 245. CrossRef - Tannin extracted from Penthorum chinense Pursh, a potential drug with antimicrobial and antibiofilm effects against methicillin-sensitive Staphylococcus aureus and methicillin-resistant Staphylococcus aureus
Junyuan Qin, Lei Yu, Fu Peng, Xin Ye, Gangmin Li, Chen Sun, Fang Cheng, Cheng Peng, Xiaofang Xie
Frontiers in Microbiology.2023;[Epub] CrossRef - Biofilm Formation of Staphylococcus aureus from Pets, Livestock, and Wild Animals: Relationship with Clonal Lineages and Antimicrobial Resistance
Vanessa Silva, Elisete Correia, José Eduardo Pereira, Camino González-Machado, Rosa Capita, Carlos Alonso-Calleja, Gilberto Igrejas, Patrícia Poeta
Antibiotics.2022; 11(6): 772. CrossRef - Sequence Type 5 (ST5) as a Possible Predictor of Bacterial Persistence in Adult Patients with Methicillin-Resistant Staphylococcus aureus Pneumonia Treated with Vancomycin
Ya-Xin Fan, Meng-Ting Chen, Nan-Yang Li, Xiao-Fen Liu, Min-Jie Yang, Yuan-Cheng Chen, Xiao-Yu Liang, Ju-Fang Wu, Bei-Ning Guo, Si-Chao Song, Yong-Qiang Zhu, Feng-Ying Zhang, Jing-Qing Hang, Sheng-Bin Wu, Bo Shen, Hua-Yin Li, Qin Wang, Xu-Ming Luo, Qing-Ge
Microbiology Spectrum.2022;[Epub] CrossRef - Ten-step asymmetric total syntheses of potent antibiotics anthracimycin and anthracimycin B
Peilin Tian, Wenkang Ye, Xiayan Zhang, Yi Tong, Pei-Yuan Qian, Rongbiao Tong
Chemical Science.2022; 13(43): 12776. CrossRef - Biofilm Formation and Prevalence of Biofilm-Related Genes Among Clinical Strains of Multidrug-Resistant Staphylococcus aureus
Natalia Kaźmierczak, Bartłomiej Grygorcewicz, Lidia Piechowicz
Microbial Drug Resistance.2021; 27(7): 956. CrossRef - Correlation Between Biofilm-Formation and the Antibiotic Resistant Phenotype in Staphylococcus aureus Isolates: A Laboratory-Based Study in Hungary and a Review of the Literature
Seyyed Askhan Senobar Tahaei, Anette Stájer, Ibrahim Barrak, Eszter Ostorházi, Dóra Szabó, Márió Gajdács
Infection and Drug Resistance.2021; Volume 14: 1155. CrossRef - Investigation of biofilm formation in methicillin-resistant Staphylococcus aureus associated with bacteraemia in a tertiary hospital
Wen Kiong Niek, Cindy Shuan Ju Teh, Nuryana Idris, Kwai Lin Thong, Soo Tein Ngoi, Sasheela Sri La Sri Ponnampalavanar
Folia Microbiologica.2021; 66(5): 741. CrossRef - Biofilm Formation of Multidrug-Resistant MRSA Strains Isolated from Different Types of Human Infections
Vanessa Silva, Luciana Almeida, Vânia Gaio, Nuno Cerca, Vera Manageiro, Manuela Caniça, José L. Capelo, Gilberto Igrejas, Patrícia Poeta
Pathogens.2021; 10(8): 970. CrossRef - Biofilm properties in relation to treatment outcome in patients with first-time periprosthetic hip or knee joint infection
Karin Svensson Malchau, Jonatan Tillander, Magdalena Zaborowska, Maria Hoffman, Iñigo Lasa, Peter Thomsen, Henrik Malchau, Ola Rolfson, Margarita Trobos
Journal of Orthopaedic Translation.2021; 30: 31. CrossRef - Evaluation of antimicrobial peptide LL-37 for treatment of Staphylococcus aureus biofilm on titanium plate
Jiantong Wei, Xuepeng Cao, Jun Qian, Zhixia Liu, Xulong Wang, Qinliuye Su, Yongpin Wang, Ruimin Xie, Xiang Li
Medicine.2021; 100(44): e27426. CrossRef - Biofilm Formation and its Association with Antibiotic Susceptibility Pattern in Methicillin-resistant Staphylococcus aureus isolates
Bajarangi Lal Chaudhary, Dakshina Bisht, Sameer Singh Faujdar
Journal of Pure and Applied Microbiology.2021; 15(4): 2041. CrossRef - The Role of Staphylococcus aureus YycFG in Gene Regulation, Biofilm Organization and Drug Resistance
Shizhou Wu, Junqi Zhang, Qi Peng, Yunjie Liu, Lei Lei, Hui Zhang
Antibiotics.2021; 10(12): 1555. CrossRef - Development of the Bacterial Spectrum and Antimicrobial Resistance in Surgical Site Infections of Trauma Patients
Rico Eisner, Norman Lippmann, Christoph Josten, Arne C. Rodloff, Daniel Behrendt
Surgical Infections.2020; 21(8): 684. CrossRef - Reduction of Pseudomonas aeruginosa biofilm formation through the application of nanoscale vibration
Shaun N. Robertson, Peter G. Childs, Ayorinde Akinbobola, Fiona L. Henriquez, Gordon Ramage, Stuart Reid, William G. Mackay, Craig Williams
Journal of Bioscience and Bioengineering.2020; 129(3): 379. CrossRef - Virulence factors and antimicrobial resistance of Staphylococcus aureus isolated from the production process of Minas artisanal cheese from the region of Campo das Vertentes, Brazil
R.D. Castro, S.H.S.P. Pedroso, S.H.C. Sandes, G.O. Silva, K.C.M. Luiz, R.S. Dias, R.A.T. Filho, H.C.P. Figueiredo, S.G. Santos, A.C. Nunes, M.R. Souza
Journal of Dairy Science.2020; 103(3): 2098. CrossRef - Investigation of biofilm production and its association with genetic and phenotypic characteristics of OM (osteomyelitis) and non-OM orthopedic Staphylococcus aureus
Shengpeng Yu, Bei Jiang, Chao Jia, Hongri Wu, Jie Shen, Xiaomei Hu, Zhao Xie
Annals of Clinical Microbiology and Antimicrobials.2020;[Epub] CrossRef - Busting biofilms: free-living amoebae disrupt preformed methicillin-resistant Staphylococcus aureus (MRSA) and Mycobacterium bovis biofilms
Kevin H. Martin, Grace I. Borlee, William H. Wheat, Mary Jackson, Bradley R. Borlee
Microbiology .2020; 166(8): 695. CrossRef - Biofilm formation and molecular characterization of methicillin-resistant Staphylococcus aureus strains isolated from the patients, personnel, air and environment of ICUs
Fatemeh Tahmasbi, Raheleh Sheikhi, Ali Ashraf, Ali Mojtahedi
Gene Reports.2020; 20: 100736. CrossRef - Characteristics of Clinically Significant Invasive Staphylococcus aureus Infections in a Tertiary Care Centre
Gillaine Vail Pinto, Archana Bhat K., Sevitha Bhat
Journal of Pure and Applied Microbiology.2020; 14(2): 1487. CrossRef - Status of Biofilm-Forming Genes among Jordanian Nasal Carriers of Methicillin-Sensitive and Methicillin-Resistant Staphylococcus aureus
Ashraf I Khasawneh, Nisreen Himsawi, Jumana Abu-Raideh, Muna A. Salameh, Mohammad Al-Tamimi, Sameer Al Haj Mahmoud, Tareq Saleh
Iranian Biomedical Journal.2020; 24(6): 381. CrossRef - Detection of Heavy Metal Tolerance among different MLSB Resistance Phenotypes of Methicillin-Resistant S. aureus (MRSA)
Sara H. Mohamed, Maram M.S. Elshahed, Yasmine M. Saied, Mahmoud S.M. Mohamed, Gamal H. Osman
Journal of Pure and Applied Microbiology.2020; 14(3): 1905. CrossRef - Membrane Vesicles Are the Dominant Structural Components of Ceftazidime-Induced Biofilm Formation in an Oxacillin-Sensitive MRSA
Xinlong He, Shuang Li, Yi Yin, Jiahui Xu, Weijuan Gong, Guocai Li, Li Qian, Yinyan Yin, Xiaoqin He, Tingting Guo, Yuzheng Huang, Feng Lu, Jun Cao
Frontiers in Microbiology.2019;[Epub] CrossRef - Biofilm formation byStaphylococcus aureusclinical isolates correlates with the infection type
Jakub M. Kwiecinski, Gunnar Jacobsson, Alexander R. Horswill, Elisabet Josefsson, Tao Jin
Infectious Diseases.2019; 51(6): 446. CrossRef - Natural Products That Target Virulence Factors in Antibiotic-Resistant Staphylococcus aureus
Shuai-Cheng Wu, Fei Liu, Kui Zhu, Jian-Zhong Shen
Journal of Agricultural and Food Chemistry.2019; 67(48): 13195. CrossRef - Antimicrobial resistance and genetic characterization of coagulase-negative staphylococci from bovine mastitis milk samples in Korea
Su-Jeong Kim, Dong Chan Moon, Seung-Chun Park, Hee Young Kang, Seok Hyeon Na, Suk-Kyung Lim
Journal of Dairy Science.2019; 102(12): 11439. CrossRef - Options and Limitations in Clinical Investigation of Bacterial Biofilms
Maria Magana, Christina Sereti, Anastasios Ioannidis, Courtney A. Mitchell, Anthony R. Ball, Emmanouil Magiorkinis, Stylianos Chatzipanagiotou, Michael R. Hamblin, Maria Hadjifrangiskou, George P. Tegos
Clinical Microbiology Reviews.2018;[Epub] CrossRef - Fighting biofilms with lantibiotics and other groups of bacteriocins
Harsh Mathur, Des Field, Mary C. Rea, Paul D. Cotter, Colin Hill, R. Paul Ross
npj Biofilms and Microbiomes.2018;[Epub] CrossRef - Prevalence of Panton-Valentine Leukocidin Gene among Community AcquiredStaphylococcus aureus: A Real-Time PCR Study
Amit Karmakar, Debarati Jana, Kunal Dutta, Parimal Dua, Chandradipa Ghosh
Journal of Pathogens.2018; 2018: 1. CrossRef - Biofilm Forming Ability and Spa Gene Polymorphism in Methicillin Resistant Staphylococcus aureus Clinical Isolates in North of Iran
L. Asadpour
Molecular Genetics, Microbiology and Virology.2018; 33(1): 55. CrossRef - Clinical and Genetic Risk Factors for Biofilm-Forming Staphylococcus aureus
Megan K. Luther, Diane M. Parente, Aisling R. Caffrey, Kathryn E. Daffinee, Vrishali V. Lopes, Emily T. Martin, Kerry L. LaPlante
Antimicrobial Agents and Chemotherapy.2018;[Epub] CrossRef - Comparative study of virulence factors among methicillin resistant Staphylococcus aureus clinical isolates
Ons Haddad, Abderrahmen Merghni, Aida Elargoubi, Hajer Rhim, Yosr Kadri, Maha Mastouri
BMC Infectious Diseases.2018;[Epub] CrossRef - Microbacterium telephonicum sp. nov., isolated from the screen of a cellular phone
Praveen Rahi, Rashmi Kurli, Aabeejjeet N. Pansare, Mitesh Khairnar, Shubhangi Jagtap, Nisha B. Patel, Syed G. Dastager, Paul A. Lawson, Yogesh S. Shouche
International Journal of Systematic and Evolutiona.2018; 68(4): 1052. CrossRef - Evaluation of Biofilm Formation and Presence of <italic>Ica</italic> Genes in <italic>Staphylococcus epidermidis</italic> Clinical Isolates
Maryam Kord, Abdollah Ardebili, Maryam Jamalan, Roghaye Jahanbakhsh, Naser Behnampour, Ezzat Allah Ghaemi
Osong Public Health and Research Perspectives.2018; 9(4): 160. CrossRef - Utilization of supercritical carbon dioxide in fabrication of cellulose acetate films with anti-biofilm effects against Pseudomonas aeruginosa and Staphylococcus aureus
Irena Zizovic, Lidija Senerovic, Ivana Moric, Tijana Adamovic, Milena Jovanovic, Melina Kalagasidis Krusic, Dusan Misic, Dusica Stojanovic, Stoja Milovanovic
The Journal of Supercritical Fluids.2018; 140: 11. CrossRef - Desiccation and ethanol resistances of multidrug resistant Acinetobacter baumannii embedded in biofilm: The favorable antiseptic efficacy of combination chlorhexidine gluconate and ethanol
Shyh-Ren Chiang, Fang Jung, Hung-Jen Tang, Chung-Hua Chen, Chi-Chung Chen, Hsiu-Yin Chou, Yin-Ching Chuang
Journal of Microbiology, Immunology and Infection.2018; 51(6): 770. CrossRef - Biofilm Formation and Its Relationship with the Molecular Characteristics of Food‐Related Methicillin‐Resistant Staphylococcus aureus (MRSA)
Alberto Vergara, Giovanni Normanno, Pierluigi Di Ciccio, Francesca Pedonese, Roberta Nuvoloni, Antonio Parisi, Gianfranco Santagada, Angelo Colagiorgi, Emanuela Zanardi, Sergio Ghidini, Adriana Ianieri
Journal of Food Science.2017; 82(10): 2364. CrossRef - MRSA decolonization failure—are biofilms the missing link?
Frank Günther, Brigitte Blessing, Evelina Tacconelli, Nico T. Mutters
Antimicrobial Resistance & Infection Control.2017;[Epub] CrossRef - The role of biofilms in persistent infections and factors involved inica-independent biofilm development and gene regulation inStaphylococcus aureus
Agnes Marie Sá Figueiredo, Fabienne Antunes Ferreira, Cristiana Ossaille Beltrame, Marina Farrel Côrtes
Critical Reviews in Microbiology.2017; 43(5): 602. CrossRef - Chlorhexidine whole-body washing of patients reduces methicillin-resistant Staphylococcus aureus and has a direct effect on the distribution of the ST5-MRSA-II (New York/Japan) clone
Maria Elena Velázquez-Meza, Soraya Mendoza-Olazarán, Gabriela Echániz-Aviles, Adrián Camacho-Ortiz, Michel Fernando Martínez-Reséndez, Vanessa Valero-Moreno, Elvira Garza-González
Journal of Medical Microbiology.2017; 66(6): 721. CrossRef - Bovine origin Staphylococcus aureus: A new zoonotic agent?
Relangi Tulasi Rao, Kannan Jayakumar, Pavitra Kumar
Veterinary World.2017; 10(10): 1275. CrossRef - Affinity interactions drive post-implantation drug filling, even in the presence of bacterial biofilm
Erika L. Cyphert, Sean T. Zuckerman, Julius N. Korley, Horst A. von Recum
Acta Biomaterialia.2017; 57: 95. CrossRef - Emerging technologies for long-term antimicrobial device coatings: advantages and limitations
Erika L Cyphert, Horst A von Recum
Experimental Biology and Medicine.2017; 242(8): 788. CrossRef - Methicillin-resistant food-related Staphylococcus aureus: a review of current knowledge and biofilm formation for future studies and applications
Agapi I. Doulgeraki, Pierluigi Di Ciccio, Adriana Ianieri, George-John E. Nychas
Research in Microbiology.2017; 168(1): 1. CrossRef - Biofilm formation of Brazilian meticillin-resistant Staphylococcus aureus strains: prevalence of biofilm determinants and clonal profiles
Deivid William da Fonseca Batistão, Paola Amaral de Campos, Nayara Caroline Camilo, Sabrina Royer, Bruna Fuga Araújo, Karinne Spirandelli Carvalho Naves, Margarida Martins, Maria Olívia Pereira, Mariana Henriques, Paulo Pinto Gontijo-Filho, Cláudia Botelh
Journal of Medical Microbiology.2016; 65(4): 286. CrossRef - Efficacy of Linezolid and Fosfomycin in Catheter-Related Biofilm Infection Caused by Methicillin-ResistantStaphylococcus aureus
Dong Chai, Xu Liu, Rui Wang, Yan Bai, Yun Cai
BioMed Research International.2016; 2016: 1. CrossRef - Methicillin-resistant Staphylococcus aureus biofilm formation on dacryocystorhinostomy silicone tubes depends on the genetic lineage
Ivana Ćirković, Miroslav Knežević, Dragana D. Božić, Dejan Rašić, Anders Rhod Larsen, Slobodanka Đukić
Graefe's Archive for Clinical and Experimental Oph.2015; 253(1): 77. CrossRef - Prevalence and Antimicrobial-Resistance of Staphylococcus aureus Isolated from Blood Culture in University Hospital, Turkey
K Murat
Global Journal of Infectious Diseases and Clinical.2015; : 010. CrossRef


 PubReader
PubReader Cite
Cite
