Search
- Page Path
- HOME > Search
Original Article
- Phylogenetic and genome-wide mutational analysis of SARS-CoV-2 strains circulating in Nigeria: no implications for attenuated COVID-19 outcomes
- Daniel B. Kolawole, Malachy I. Okeke
- Osong Public Health Res Perspect. 2022;13(2):101-113. Published online April 22, 2022
- DOI: https://doi.org/10.24171/j.phrp.2021.0329
- 3,295 View
- 64 Download
-
 Graphical Abstract
Graphical Abstract
 Abstract
Abstract
 PDF
PDF 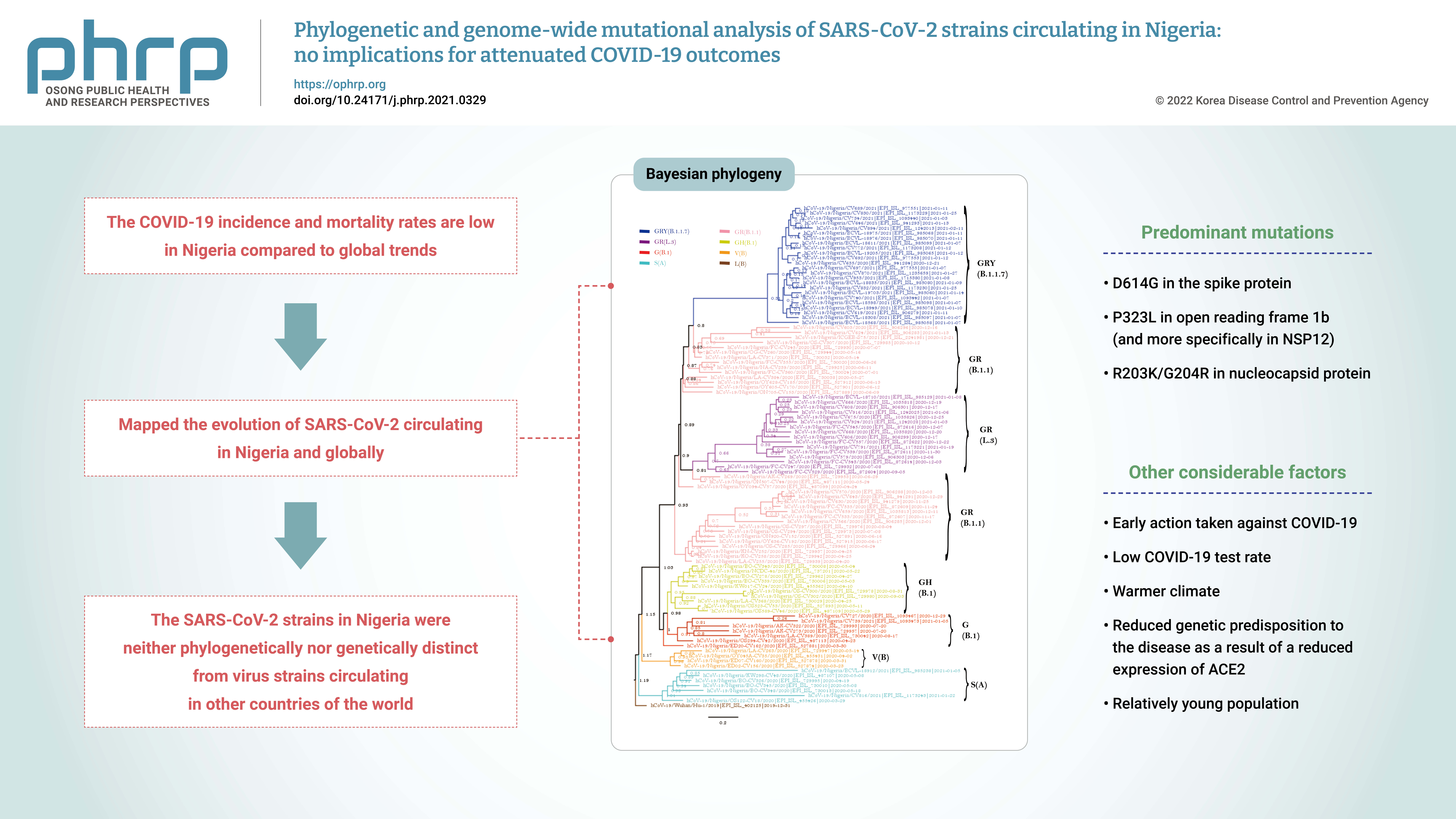
- Objectives
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is the causative agent of coronavirus disease 2019 (COVID-19). The COVID-19 incidence and mortality rates are low in Nigeria compared to global trends. This research mapped the evolution of SARS-CoV-2 circulating in Nigeria and globally to determine whether the Nigerian isolates are genetically distinct from strains circulating in regions of the world with a high disease burden. Methods: Bayesian phylogenetics using BEAST 2.0, genetic similarity analyses, and genomewide mutational analyses were used to characterize the strains of SARS-CoV-2 isolated in Nigeria. Results: SARS-CoV-2 strains isolated in Nigeria showed multiple lineages and possible introductions from Europe and Asia. Phylogenetic clustering and sequence similarity analyses demonstrated that Nigerian isolates were not genetically distinct from strains isolated in other parts of the globe. Mutational analysis demonstrated that the D614G mutation in the spike protein, the P323L mutation in open reading frame 1b (and more specifically in NSP12), and the R203K/ G204R mutation pair in the nucleocapsid protein were most prevalent in the Nigerian isolates. Conclusion: The SARS-CoV-2 strains in Nigeria were neither phylogenetically nor genetically distinct from virus strains circulating in other countries of the world. Thus, differences in SARS-CoV-2 genomes are not a plausible explanation for the attenuated COVID-19 outcomes in Nigeria.
Article
- Phylogenetic Analysis of the Rotavirus Genotypes Originated from Children < 5 Years of Age in 16 Cities in South Korea, between 2000 and 2004
- Ho-Kyung Oh, Seung-Hwa Hong, Byung-Yoon Ahn, Hye-Kyoung Min
- Osong Public Health Res Perspect. 2012;3(1):36-42. Published online December 31, 2011
- DOI: https://doi.org/10.1016/j.phrp.2012.01.006
- 2,750 View
- 16 Download
- 6 Crossref
-
 Abstract
Abstract
 PDF
PDF - Objectives
The purpose of this study was to examine the diversity of the G and P types of human rotavirus strains isolated in South Korea during 2000 to 2004.
Methods
We selected 38 Group A rotavirus isolates among 652 fecal samples, which were collected from infants and children < 5 years of age with acute gastroenteritis or diarrhea admitted in 8 hospitals representative of five provinces of South Korea between 2000 and 2004. Rotavirus P- and G-genotypes were determined by nucleotide sequencing and phylogenetic analysis was performed.
Results
One G1P[4] consisted G1-Id-P[4]-V; one G1P[6] consisted G1-Id-P[6]-Ia; nine G1P[8] consisted G1-Ib-P[8]-Ia (n=3), G1-Ic-P[8]-Ia (n=1), and G1-Id-P[8]-Ia (n=5); 13 G2P[4] consisted G2-V-P[4]-V; two G3P[4] consisted G3-IIId-P[4]-V; five G3P[8] consisted G3-IIId-P[8]-Ia; four G4P[6] consisted G4-Ie-P[6]-Ia; two G4P[8] consisted G4-Ie-P[8]-II; one G9P[6] consisted G9-III-P[6]-Ia.
Conclusions
A considerable amount of rotavirus genotypic diversity was detected in South Korea from 2000 to 2004. These findings are important to develop the effective vaccines and to undertake epidemiologic studies. -
Citations
Citations to this article as recorded by- Long-term monitoring of G1P[8] rotaviruses circulating without vaccine pressure in Nizhny Novgorod, Russia, 1984-2019
N. A. Novikova, T. A. Sashina, N. V. Epifanova, A. U. Kashnikov, O. V. Morozova
Archives of Virology.2020; 165(4): 865. CrossRef - Prevalence, risk factors, and clinical characteristics of rotavirus and adenovirus among Lebanese hospitalized children with acute gastroenteritis
Rasha Zaraket, Ali Salami, Marwan Bahmad, Ali El Roz, Batoul Khalaf, Ghassan Ghssein, Hisham F. Bahmad
Heliyon.2020; 6(6): e04248. CrossRef - Prevalence and Genotypic Distribution of Rotavirus in Thailand: A Multicenter Study
Pimmada Sakpaisal, Sasikorn Silapong, Amara Yowang, Gaysorn Boonyasakyothin, Boonyaorn Yuttayong, Umaporn Suksawad, Siriporn Sornsakrin, Paphavee Lertsethtakarn, Ladaporn Bodhidatta, John M. Crawford, Carl J. Mason
The American Journal of Tropical Medicine and Hygi.2019; 100(5): 1258. CrossRef - Monitoring Shedding of Five Genotypes of RotaTeq Vaccine Viruses by Genotype-Specific Real-Time Reverse Transcription-PCR Assays
Yuki Higashimoto, Masaru Ihira, Yu Miyazaki, Ayumi Kuboshiki, Sayaka Yoshinaga, Hiroyuki Hiramatsu, Ryota Suzuki, Masafumi Miyata, Hiroki Miura, Satoshi Komoto, Jun Yukitake, Koki Taniguchi, Yoshiki Kawamura, Tetsushi Yoshikawa, Yi-Wei Tang
Journal of Clinical Microbiology.2018;[Epub] CrossRef - Molecular analysis of group A rotaviruses detected in hospitalized children from Rawalpindi, Pakistan during 2014
Massab Umair, Bilal Haider Abbasi, Nadia Nisar, Muhammad Masroor Alam, Salmaan Sharif, Shahzad Shaukat, Muhammad Suleman Rana, Adnan Khurshid, Ghulam Mujtaba, Uzma Bashir Aamir, Syed Sohail Zahoor Zaidi
Infection, Genetics and Evolution.2017; 53: 160. CrossRef - Analysis of rotavirus genotypes in Korea during 2013: An increase in the G2P[4] genotype after the introduction of rotavirus vaccines
Jae-Seok Kim, Hyun Soo Kim, Jungwon Hyun, Han-Sung Kim, Wonkeun Song, Kyu Man Lee, Seon-Hee Shin
Vaccine.2014; 32(48): 6396. CrossRef
- Long-term monitoring of G1P[8] rotaviruses circulating without vaccine pressure in Nizhny Novgorod, Russia, 1984-2019



 First
First Prev
Prev


