The Prevalence of CYP2B6 Gene Polymorphisms in Malaria-endemic Population of Timor in East Nusa Tenggara Indonesia
Article information
Abstract
Objectives
The CYP2B6 is one of the most polymorphic CYP genes in humans that has the potential to modify the pharmacological and toxicological responses to clinically important drugs such as antimalarial artemisinin and its derivatives. The aim of the study was to determine the frequency of CYP2B6 polymorphisms in Timor malaria endemic area, East Nusa Tenggara, Indonesia where Artemisin-based Combination Therapy (ACT) has been used to treat uncomplicated malaria.
Methods
A total of 109 healthy subjects were participated in this study. CYP2B6*4, *6 and *9 polymorphisms were analyzed using PCR-RFLP to confirm the SNPs prevalence of 516G>T and 785A>G in exon 4 and 5.
Results
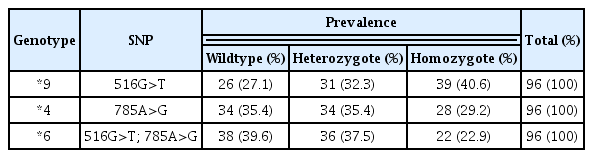
There were 96 subjects included in the analysis. In the exon 4 of CYP2B6 516G>T, the frequency of the T mutation was 37.5% (39/96), and the wildtype 27.1% (26/96). In the exon 5, CYP2B6 785A>G mutant was detected in 29.2% (28/96) of individuals, and the wildtype allele in 35.4% (34/96). The frequency of CYP2B6*9 (516G>T), CYP2B6*4 (785A>G) and CYP2B6*6 (516G>T and 785A>G) were 40.6%, 29.2% and 22.9%, respectively. The prevalence of these CYP2B6 gene polymorphisms in Timorian ethnic were higher than that in Malay, Han Chinese, Indian, and Egyptian populations.
Conclusion
The prevalence of these CYP2B6 516G>T and 785A>G polymorphisms in Timorian ethnic is higher than that in other populations. These polymorphisms may affect the metabolism of artemisinin and its derivatives.
Introduction
Cytochrome P450 2B6 (CYP2B6) is a significant component of the cytochrome P450 enzyme system, with an average relative abundance of 6% of the total hepatic cytochrome P450 content. This enzyme plays an important role in the biotransformation of drugs and other xenobiotics. The CYP2B6 mediates the metabolic activation and inactivation of various clinically important drugs, including antiretrovirals nevirpine and efavirenz, anticancer cyclophosphamide, anesthetic propofol and antimalarial drugs artemisinin and its derivatives [1,2]. The CYP2B6 is one of the most polymorphic CYP genes in humans and these polymorphisms have been demonstrated to affect the pharmacological and toxicological responses to clinically important drugs. Currently, 37 distinct star-alleles, over 50 determined gene haplotypes, and more than 100 described single-nucleotide polymorphisms (SNPs) of the CYP2B6 genes have been reported and are listed on the cypalleles websites [3,4].
Some of the CYP2B6 variant alleles occur with relatively high frequencies across different population are CYP2B6*6, CYP2B6*4 and CYP2B6*9 alleles [4,5]. The CYP2B6*6 [516G>T (Q172H) and 785A>G (K262R)] is the most common allele in all populations which occur in about 15 to over 60% in different population [4,6]. This CYP2B6*6 genotype is associated with decreased protein expression lead to decrease or increase activities depending on its drug substrates [2,7]. The CYP2B6*4 [785A>G (K262R)] is another important allele which occurs in about 9 to 63% depending on the ethnicity of the population studied [8,9]. This CYP2B6*4 genotype is associated with increased protein expression and CYP2B6 activity [3,10]. Whereas the CYP2B6*9 [516G>T (Q172H)] occurs in about 0 to 35.7% in different population [6]. This CYP2B6*9 [516G>T (Q172H)] is associated with poor metabolizer and more side effects of efavirenz.
Malaria is a major public health problem in most tropical countries, including Indonesia. Although National Malaria Control Program in Indonesia has made progress to reduce Annual Parasite Incidence (API) from 1.75 in 2011 to 0.85 in 2015, malaria is still endemic in East Nusa Tenggara with API of 7 [11]. Artemisin-based Combination Therapy (ACT) has been used in the uncomplicated malaria treatment in endemic area in Indonesia, including in East Nusa Tenggara. Some artemisinin derivatives have been shown to be metabolized by polymorphic CYP2B6. The aim of the study was to determine the frequency of CYP2B6 (516G>T and 785A>G) polymorphisms in Timorian ethnic, East Nusa Tenggara, Indonesia. The study concerning CYP2B6 polymorphisms has not been reported, yet. Such a data obtained in this study could provide valuable information on clinical responses of artemisinin derivatives and further would be helpful for personalized medicine.
Materials and Methods
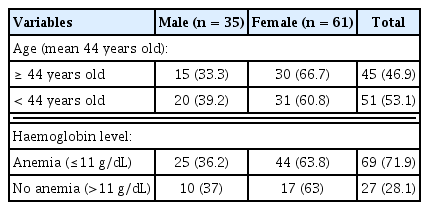
This was a cross sectional study conducted during four-month period from August to December 2017 to determine frequency of CYP2B6 (516G>T and 785A>G) polymorphisms of Timorian ethnic in malaria endemic area Amanatun Selatan Sub-district, East Nusa Tenggara Province, Indonesia. In total, 109 healthy volunteers with the aged 14 years or older and haemoglobin concentrations ≥ 11 g/dL participated in the study after completed signed informed consent forms. The protocol of the study was approved by the Ethic Research Committee from School of Medicine, Atma Jaya Catholic University of Indonesia (Ref no.: 07/03/FK/KEP/2016).
Approximately 1–2 mL of whole blood sample for each participant was collected using EDTA anticoagulant tubes (BD vacutainer 5 mL). Samples were transported to School of Medicine, Atma Jaya Catholic University of Indonesia and stored at −20°C prior to analysis. Genomic DNA was extracted from whole blood following the manufacturer’s protocols (Promega, Madison, USA). DNA concentrations was determined using a nanodrop spectrophotometer and purified DNA was stored at −20°C.
Polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP) was used for genotyping CYP2B6 in both exon 4 and 5. Specific primers were designed for the PCR-RFLP: F (5’-GGT-CTG-CCC-ATC-TAT-AAA-C-3’) and R (5’-CTC-CCT-CTG-TCT-TTC-ATT-GTG-T-3’), and for 785A>G primer F (5’-GAC-AGA-AGG-ATG-AGG-GAG-GAA-3’) and R (5’-CTC-CCT-CTG-TCT-TTC-ATT-CTG-3’) [12]. The PCR-RFLP was identified using Sty1 restriction enzyme mapping for A>G giving 468, 27TA9, 171, and 161 base pair (bp) fragments. The restriction enzyme Bsr1 was used to identify G>T, giving restriction fragments of 509, 268, and 241bp. Allele and genotype frequencies for the SNPs investigated were determined by direct counting. Deviation from Hardy-Weiberg equilibrium expectations were determined using the chi-square test. The data were shown in the distribution prevalence in the form of tables with a confidence interval (CI) of 95%.
Results
A total of 109 healthy subjects were participated in this study. The characteristics of subjects are presented in Table 1. PCR-RFLP genotyping was completed for 96 out of 109 (88.1%) individuals; 36.4% (35/96) males and 63.5% (61/96) females. Figure 1 shows the restriction fragments after digestion with Sty1 and Bsr1 restriction enzymes.
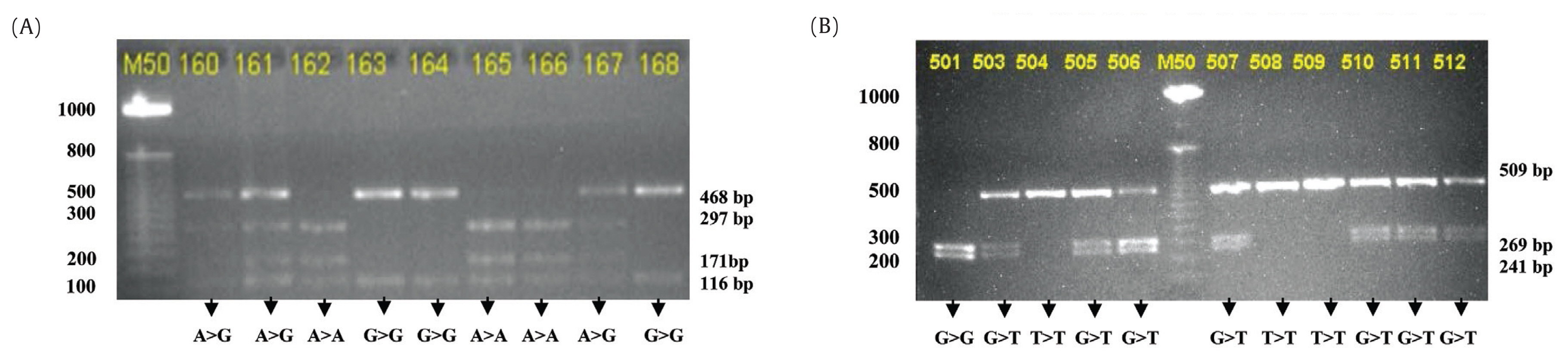
(A) Restriction fragments after digestion with Sty1 restriction enzyme to identify CYP2B6*4 (785A>G) and (B) Showing the restriction fragments after digestion with Bsr1 restriction enzyme to identify CYP2B6*9 (516G>T).
The prevalence of CYP2B6 polymorphisms was higher in exon-4 (40.6%) than in exon-5 (29.2%) as shown in Table 2.
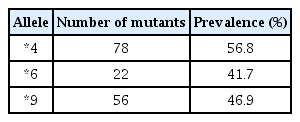
The PCR-RFLP analysis identified SNPs in both exon 4 and 5, including allele *6. There were 41.7% people with the *6 mutation (Table 3).
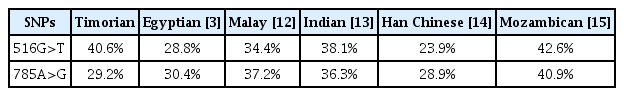
The prevalence of CYP2B6 516G>T and 785A>G polymorphisms in Timorian ethnic observed in this study was higher than that previous study in other populations, except in Mozambican population (Table 4).
Discussion
The PCR-RFLP analysis examined the prevalence of the CYP2B6 516G>T and 785A>G polymorphisms in Timorian ethnic in Amanantun Selatan Sub-district, East Nusa Tenggara, Indonesia. The prevalence of CYP2B6 516G>T and CYP2B6 785A>G were 40.6% and 29.2%, respectively. The prevalence of both alleles in Timorian ethnic was higher than that in other populations, such as Malays, Chinese, Indian, and Egyptian and except in Mozambican [3,12–15]. No significant difference was observed between the prevalence of elleles using Hardy Weinberg equation analysis. The prevalence of the CYP2B6 variants genotype in Timorian ethnic was similar to the Papua New Guinean’s (the PNGs) and comparable to the other populations. In both populations, the highest genotype variant was *6/*6 (homozygous mutant), followed by *1/*6 (heterozygous). The prevalence of the *6/*6 and *1/*6 genotype variants in the Timorian ethnic was 22.9% and 37.5%, respectively, while in PNGs it was 43% and 36%, respectively. The frequency of allele *6 in Timorian ethnic was 41.7% also similar to that in PNG (62%). This pattern might be explained by the similar ancestry of the Timorian ethnic, who are descended from Austronesians and Melanesians, with the PNGs also descendants of the Melanesians [16]. In the Negroid, Mongoloid and Caucasian population, such as West African, Chinese, Japanese, Caucasian-American, Hispanic-American, and South Iranian, the prevalence of *1/*6 were higher than the other variants (except the wild type). Because this study used the PCR-RFLP method, the CYP2B6 genotype variants *1/*6 and *4/*9 showed similar patterns on DNA visualization. The data in Table 4 assumed that the *1/*6 allele was shown by both the 516G>T and 785A>G mutations. For more precise results, DNA genotyping must be performed [17].
In previous studies, the prevalence of CYP2B6*6 was found to be higher than CYP2B6*4 and CYP2B6*9 in all populations studied. In the Timorian ethnic, the prevalence of CYP2B6*6, CYP2B6*4, and CYP2B6*9 were 22.9%, 29.2%, and 40.7%, respectively. The prevalence of CYP2B6*6 in the Timorian ethnic was higher than that in Korean (15.9%), Japanese (17.6%), Chinese (18.4%), Mongolian (21%), and was similar with Southeast Iranian (23.1%), Caucasian-American (28%), Hispanic-American (30%), and African-American populations (34%). As previously described, the similarity of the prevalence of CYP2B6*6 in Timorian ethnic and West African populations was surprising considering the difference in their genotype variant. The prevalence of CYP2B6*9 was highest in the Timorian ethnic (40.7%) compared to other populations [17].
Individuals with the CYP2B6*6 gene metabolizes drugs slower than individuals with the CYP2B6*1 gene, because CYP2B6*6 is considered a slow metabolizer [18]. When these individuals are given drugs in the prodrug, the effect of the drug is decreased causing therapy failure and prodrug accumulation in the blood. This condition may increase the side effects of the drugs. When the drug is given in the active form, the secretion rate decreases and the concentration of active drug in the blood increases along with potential side effects, therefore, lower doses may be required in these individuals to reduce the side effects. Artemisinin and its derivatives are frequently used in East Timor for the treatment of uncomplicated malaria. The high prevalence of CYP2B6*6 in the Timor ethnic should be considered when giving these antimalarial drugs because of the slow metabolizer characteristic. Artemisinins are the newest class of antimalarial agents. They are generally well tolerated and as the WHO recommended, oral ACT, such as dihydroartemisinin–piperaquine, artemether–lumefantrine, artesunate–mefloquine, and artesunate–amodiaquine have rapidly become first-line drugs for the treatment of uncomplicated malaria in endemic countries. The metabolism of artemisinins-based compounds is complex. Artesunate, artemether, and arteether are primarily metabolized by CYP3A4, CYP3A5, and CYP2A6 with a minor contribution from CYP2B6 to form dihydroartemisinin, which is subsequently inactivated via UGT1A9 and UGT2B7. The artemisinin, on the other hand, is primarily metabolized by CYP2B6 with a minor contribution from CYP3A4 and CYP2A6. Artemether, artemisinin, and dihydroartemisinin have all been shown to induce CYP3A4, CYP2B6, and ABCB1 through activation of pregnane X-receptor and constitutive androstane receptor [18,4]. Until now, pharmacogenetic data on artemisinin compounds is very limited. Studies the effect of CYP2B6 gene polymorphisms on the pharmacological and toxicological responses to artemisinin and its derivatives are needed.
Conclusion
The prevalence of CYP2B6 polymorphisms was determined in malaria-endemic Timor population in East Nusa Tenggara, Indonesia. The frequency of CYP2B6*9 (516G>T), CYP2B6*4 (785A>G) and CYP2B6*6 (516G>T and 785A>G) are 40.6%, 29.2% and 22.9%, respectively. The prevalence of the CYP2B6 gene polymorphism is higher than that in Malay, Han Chinese, Indian, and Egyptian populations. This polymorphism may affect the metabolism of artemisinin and its derivatives. Further study will be performed to investigate the effect of the CYP2B6 polymorphisms to pharmacokinetics of artemisinin and its derivatives.
Acknowledgements
Authors would like to thanks all volunteers from the Amanatun Selatan Sub-district, South Central Timor District who participated in this study. This study was financial supported by the Ministry of Research, Technology and Higher Education of Republic of Indonesia and School of Medicine, Atma Jaya Catholic University of Indonesia through Doctoral Dissertation Grant in 2017 (Grant no.: 1074/III/LPPM-HD/05/2017). We would also like to thank all technicians from the Biomedical Laboratory, School of Medicine, Atma Jaya Catholic University and our colleagues from Universitas Gadjah Mada, Yogyakarta for the valuable assistances during the study.
Notes
Conflicts of Interest
There was no potential conflicts of interest relevant to this article.