Identification of Coronavirus Isolated from a Patient in Korea with COVID-19
Article information
Abstract
Objectives
Following reports of patients with unexplained pneumonia at the end of December 2019 in Wuhan, China, the causative agent was identified as coronavirus (SARS-CoV-2), and the 2019 novel coronavirus disease was named COVID-19 by the World Health Organization. Putative patients with COVID-19 have been identified in South Korea, and attempts have been made to isolate the pathogen from these patients.
Methods
Upper and lower respiratory tract secretion samples from putative patients with COVID-19 were inoculated onto cells to isolate the virus. Full genome sequencing and electron microscopy were used to identify the virus.
Results
The virus replicated in Vero cells and cytopathic effects were observed. Full genome sequencing showed that the virus genome exhibited sequence homology of more than 99.9% with SARS-CoV-2 which was isolated from patients from other countries, for instance China. Sequence homology of SARS-CoV-2 with SARS-CoV, and MERS-CoV was 77.5% and 50%, respectively. Coronavirus-specific morphology was observed by electron microscopy in virus-infected Vero cells.
Conclusion
SARS-CoV-2 was isolated from putative patients with unexplained pneumonia and intermittent coughing and fever. The isolated virus was named BetaCoV/Korea/KCDC03/2020.
Introduction
Coronavirus is an RNA virus consisting of positive-sense single-stranded RNA of approximately 27–32 kb. Coronavirus belong to the family Coronaviridae, which comprises of alpha, beta, delta, and gamma coronaviruses [1,2]. As the name indicates, the spherical external spike protein displays a characteristic crown shape when observed under an electron microscope [3,4]. The virus is known to infect a wide range of hosts including humans, other mammals, and birds. Infected hosts exhibit different clinical courses, ranging from asymptomatic to severe symptoms in their respiratory, digestive, and genital organs [1,2]. There are 6 known coronaviruses that typically cause infection in humans. Among these, coronavirus 229E, OC43, NL63, and HKU1 generally cause mild cold-like symptoms, whereas severe acute respiratory syndrome-coronavirus (SARS-CoV) in 2003, and Middle East respiratory syndrome-coronavirus (MERS-CoV) in 2012, caused severe respiratory diseases such as pneumonia and death [5,6].
Following the first outbreaks of unexplained pneumonia in Wuhan, China, in late 2019, a new coronavirus was identified as the causative agent in January 2020 [7]. As of February 11th, 2020, a total of 45,000 cases of pneumonia, have been reported from 26 countries, including China. Approximately 96.8% of all cases have been reported in China, and patients in the Hubei province account for 75% of all cases [8–10]. In Korea, the first case was reported on January 20, 2020, when SARS-CoV-2 was detected in a traveler entering Korea from Wuhan, China [11].
This study reports the full genome sequencing of SARS-CoV-2 isolated from putative the 2019 novel coronavirus disease (COVID-19) patients in Korea, by cell culture. The isolated SARS-CoV-2 was named BetaCoV/Korea/KCDC03/2020.
Materials and Methods
1. Clinical specimens and RNA extraction
Nasopharyngeal and oropharyngeal swab and sputum samples were collected from symptomatic patients to detect SARS-CoV-2 by real-time reverse transcriptase (RT)-PCR. RNA was extracted from clinical samples with a QIAamp viral RNA mini kit (QIAGEN, Hilden, Germany) following the manufacturer’s instructions. All specimens were handled under a biosafety cabinet according to laboratory biosafety guidelines of Korea Centers for Disease Control and Prevention for COVID-19.
2. Real-time RT-PCR
The optimal concentration of primers and probes, which were synthesized using a published sequence [12], was determined with the RNA transcripts of SARS-CoV. The primer and probe sequences used for RNA-dependent RNA polymerase gene detection were: 5′-GTGARATGGTCATGTGTGGCGG-3′ (Forward), 5′-CARATGTTAAASACACTATTAGCATA-3′ (Reverse) and 5′-CAGGTGGAACCTCATCAGGAGATGC-3′ (Probe in 5-FAM/3′-BHQ format) and the primer and probe sequences used for E gene detection were: 5′-ACAGGTACGTTAATAGTTAATAGCGT-3′ (Forward), 5′-ATATTGCAGCAGTACGCACACA-3′ (Reverse) and 5′-ACACTAGCCATCCTTACTGCGCTTCG-3′ (Probe in 5-FAM/3′-BHQ format). A 25-μL reaction was setup that contained 5 μL of RNA, 12.5 μL of 2 × reaction buffer provided with the Agpath IDTM 1 step RT-PCR system (Thermo Fisher Scientific, Waltham, USA), 1 μL of 25 × enzyme mixture, 1 μL of forward and reverse primers at 10 pM, and 0.5 μL of each probe at 10 pM. Reverse transcription was performed at 50°C for 30 minutes, followed by inactivation of the reverse transcriptase at 95°C for 10 minutes. PCR amplification was performed with 40 cycles at 95°C for 15 seconds and 60°C for 1 minute using an ABI 7500 Fast instrument (Thermo Fisher Scientific).
3. Virus isolation
The virus was isolated from nasopharyngeal and oropharyngeal samples from putative COVID-19 patients. Oropharyngeal samples were diluted with viral transfer medium containing nasopharyngeal swabs and antibiotics (Nystadin, penicillin-streptomycin 1:1 dilution) at 1:4 ratio and incubated for 1 hour at 4°C, before being inoculated onto Vero cells. Inoculated Vero cells were cultured at 37°C, 5% CO2 in 1× Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 2% fetal bovine serum and penicillin-streptomycin. Virus replication and isolation were confirmed through cytopathic effects, gene detection, and electron microscopy. Viral culture of SARS-CoV-2 was conducted in a biosafety Level-3 facility according to laboratory biosafety guidelines of Korea Centers for Disease Control and Prevention.
4. Next generation sequencing of viral full-length genome
Using reverse transcriptase, cDNA was synthesized from RNA extracted from the cultured cell medium in which the virus was replicated. A next generation sequencing (NGS) library was constructed after amplifying the full-length genes of the isolates using the synthesized cDNA and primers designed based on published SARS-CoV-2 DNA sequence. The prepared library was purified and analyzed with Miseq 150 PE. De novo assembly was performed on the sequenced product using Megahit to secure a full-length genome.
5. Sequencing analysis
Gene sequencing was performed using CLC Main Workbench 7.9.1. Alignment was conducted using human and animal coronavirus sequences registered in Global Initiative on Sharing All Influenza Data (GISAID) and NCBI GenBank. The phylogenetic tree was analyzed using MEGA6 with the neighbor-joining method, maximum composite likelihood-parameter distance matrix, and bootstrap values of 1,000 replicates.
6. Transmission electron microscopy
For transmission electron microscopy, the inoculated cells were prefixed by incubating in 2% paraformaldehyde and 2.5% glutaraldehyde in 0.1 M phosphate buffer (pH 7.4) to prevent the autolysis of the cells infected with virus. To minimize the chemical reaction between pre- and post-fixation, the slides were washed 3 times using the same buffer as in the fixative solution and post-fixed with 1% osmium tetroxide. After washing 3 times with deionized water, en bloc staining was performed using 0.5% uranyl acetate. Thereafter, 30%, 50%, 70%, 80%, 90%, and 100% ethanol were used sequentially in ascending concentration for dehydration, which was substituted with propylene oxide. The slides were then embedded in Epon812 plastic resin, and polymerized at 70°C for 48 hours. The prepared plastic block was cut to 70-nm thick sections using an ultramicrotome and mounted on a 100-mesh nickel grid, and electrostained with 5% uranyl acetate. The sections were observed with a transmission electron microscope (Libra120, Carl Zeiss, Germany) at an acceleration voltage of 120 kV [13–15].
Results
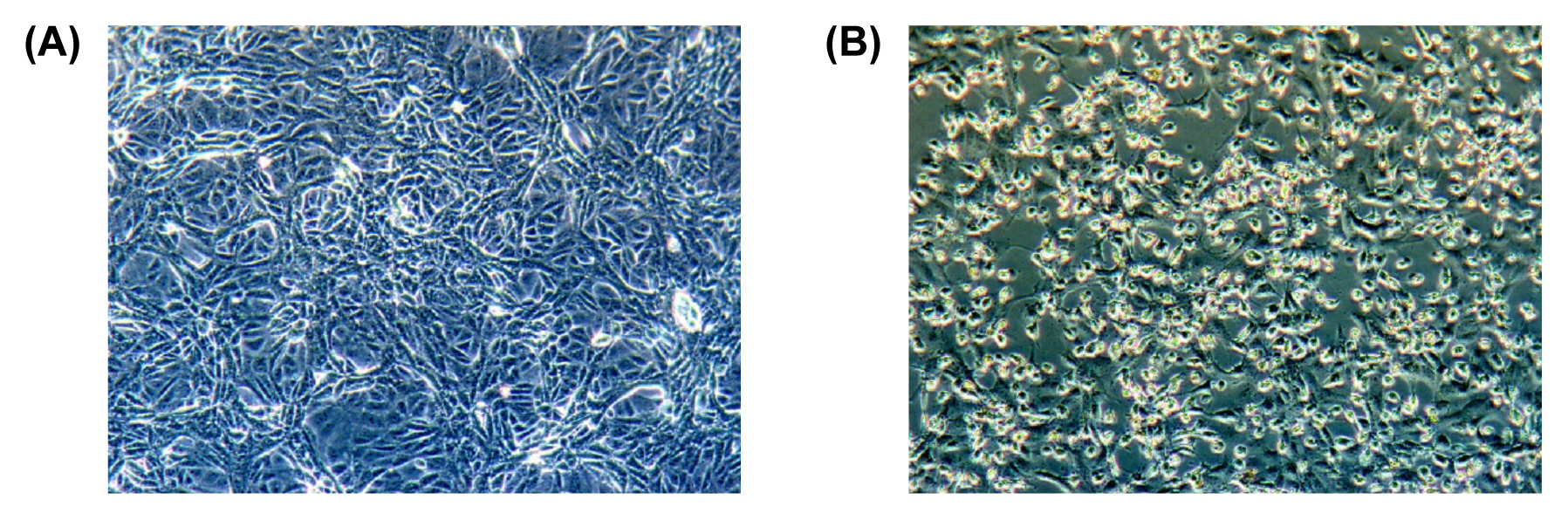
1. Virus isolation from Vero cells
Following inoculation of Vero cells with the nasopharyngeal and oropharyngeal samples, they were observed at 24-hour intervals, and the cytopathic effects were observed from 3 days after inoculation (Figure 1). The inoculated cells were harvested on the 4th day when more than 80% of the cells exhibited cytopathic effects. Virus replication was confirmed using real-time RT-PCR with RNA extracted from the cell culture medium. The Ct values were 14.40 and 18.26 for the nasopharyngeal and oropharyngeal samples, respectively, which were lower than the cycle threshold (Ct) values of 20.85 and 21.85 in the pre-inoculated samples. The number of virus copies in the samples before inoculation was 7.6 × 108 and 3.9 × 108 copy/mL, respectively, and increased by 10–70-fold to 5.4 × 1010 and 4.2 × 109, respectively, in the cell culture supernatants.
2. Analysis of the structure of the virus by electron micrographs
The structure of the virus in the cytoplasm of 3-day post-inoculation cells was examined by electron microscopy (Figure 2). Coronavirus-specific morphology was observed. Virus particle size ranged from 70–90 nm and the virus was observed in a wide range of intracellular organelles, especially in vesicles.

Thin section electron micrographs of Vero cells infected with SARS-CoV-2. Electron micrographs show representative thin sections of Vero cells infected with SARS-CoV-2; cells were collected at 48 hours after infection for examination by electron microscopy. White arrows point to aggregates of assembled intracellular virions.
3. Full-length genome and phylogenetic analysis
After inoculating cells with the nasopharyngeal and oropharyngeal samples, RNA was extracted from the virus-replicated cell culture medium. The RNA was amplified with primers for full-length gene analysis, and NGS was performed using Miseq. De novo assembly of the NGS sequence secured 28,818 bp of the full-length gene. The acquired gene was compared with 57 human and animal coronaviruses, including Wuhan/IVDC-HB-01/2019(GISAID accession ID: EPI_ISL_402119~121), which was first reported in Wuhan, 54 reported full-length SARS-CoV-2 genes, Bat-SARS-like CoV, and human SARS-CoV (Figure 3). The analysis showed that the sequence was included in the same cluster as the previously reported SARS-CoV-2 sequence and showed high homology of > 99.5% with other isolated SARS-CoV-2 sequences. The virus was named BetaCoV/Korea/KCDC03/2020, and its full-length gene sequence was registered in WHO GISAID (GISAID accession ID: EPI_ISL_407193).

Phylogenetic tree analysis of SARS-CoV-2 based on full genome nucleotide sequences using the neighbor-joining tree. Values on branches are shown as percentages based on 1,000 bootstrap replicates. (A) Gene analysis of SARS-CoV-2 and other coronaviruses, (B) gene analysis of SARS-CoV-2 and BetaCoV/Korea/KCDC03/2020.
Discussion
As of February 12th, 2020, 28 cases of COVID-19 have been reported in Korea, with the first case observed in a traveler residing in Wuhan, China. The SARS-CoV-2 was isolated from a Korean patient who had self-administered antipyretics for initial symptoms such as chills and fever. The patient had experienced intermittent coughing with sputum 3 days after the administration of antipyretics. The SARS-CoV-2 could replicate in other cells (Vero E6 and Caco-II cells), in addition to Vero cells (data not shown). The first SARS-CoV-2 was successfully isolated by inoculating human airway epithelial cells with bronchoalveolar-lavage fluid samples from a patient with pneumonia [16]. Since human airway epithelial cells (because of their resemblance to pseudostratified mucociliary epithelium) require 4–6 weeks to differentiate in vivo, isolation of SARS-CoV-2 using Vero cells or Caco-II cells is more convenient. Further studies are needed to select more sensitive cell lines suitable for virus isolation from low viral load samples.
The sequence of the suspected novel coronavirus (KCDC03) was analyzed using the sequences of 54 SARS-CoV-2 including 6 human coronaviruses, Bat-CoVs, and 51 SARS-CoV-2 that have been registered in GISAID by several countries including China, and a phylogenetic tree was produced. Results show that the isolate from Korea was clustered with the new coronavirus, SARS-CoV-2 and classified as betacoronavirus. High homology (99.94%–99.99%) was confirmed with the viral sequences reported from other countries, such as those from Wuhan/IVDC-HB-01/2019(GISAID accession ID: EPI_ISL_402119). Homology with bat CoV (bat-SL-CoVZC45), SARS-CoV (AY278741) and MERS-CoV (JX869059) was 89.1%, 77.5%, and 50%, respectively. However, the lowest homology of 99.5% was observed with BetaCoV/Wuwhan/IVDCHB-04/2020, which could be because of the inaccurate sequence of IVDCHB-04, Hence, it has been excluded from the comparative analysis in other studies.
Prior to identification of SARS-CoV-2 as the causative agent of the unknown pneumonia in Wuhan, China, pan-CoV RT-PCR was being used to detect SARS-CoV-2 in Korea. The Pan-CoV RT-PCR detects all human coronaviruses and animal-derived coronaviruses (personal communication). Since the release of the SARS-CoV-2 sequence, a real-time RT-PCR method has been established in the diagnosis of COVID-19 patients. Currently, the diagnosis of COVID-19 is based on gene detection via real-time RT-PCR. With the isolation of the causative agent, development of serological tests and rapid diagnostic tests in addition to virus detection will be required.
Acknowledgments
This study was funded by Korea Centers for Disease Control and Prevention (no.: 4800-4837-301 and 4800-4834-303). The authors thank Mr. Jun Sub Kim of Korea CDC for his help in sequencing analysis.
Notes
Conflicts of Interest
The authors have no conflicts of interest to declare.