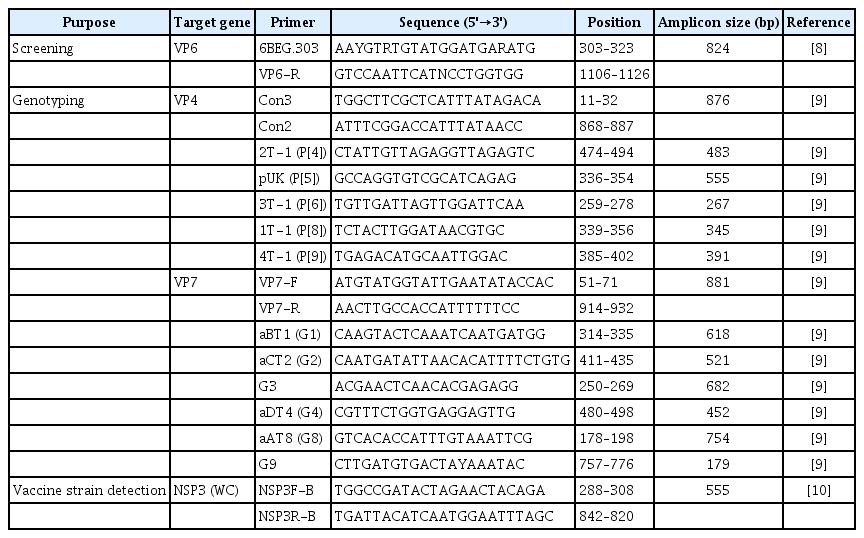
The laboratory test procedure to confirm rotavirus vaccine infection in severe complex immunodeficiency patients
Article information
Abstract
The rotavirus vaccine is a live vaccine, and there is a possibility of infection by the virus strain used in the vaccine. We investigated the process of determining whether an infection was caused by the vaccine strain in a severe complex immunodeficiency (SCID) patient with rotavirus infection. The patient was vaccinated with RotaTeq prior to being diagnosed with SCID. The testing process was conducted in the following order: confirming rotavirus infection, determining its genotype, and confirming the vaccine strain. Rotavirus infection was confirmed through enzyme immunoassay and VP6 gene detection. G1 and P[8] were identified by multiplex polymerase chain reaction for the genotype, and G3 was further identified using a single primer. By detecting the fingerprint gene (WC3) of RotaTeq, it was confirmed that the detected virus was the vaccine strain. Genotypes G1 and P[8] were identified, and the infection was suspected of having been caused by rotavirus G1P[8]. G1P[8] is the most commonly detected genotype worldwide and is not included in the recombinant strains used in vaccines. Therefore, the infection was confirmed to have been caused by the vaccine strain by analyzing the genetic relationship between VP4 and VP7. Rotavirus infection by the vaccine strain can be identified through genotyping and fingerprint gene detection. However, genetic linkage analysis will also help to identify vaccine strains.
Introduction
Rotavirus is a common cause of severe enteritis in newborns and infants worldwide. It can cause gastroenteritis with severe diarrhea and dehydration, leading to hospitalization and even death. Immunization with the rotavirus vaccine has been proven to reduce disease severity from rotavirus gastroenteritis [1–3]. Two types of rotavirus vaccines were developed in 2006 and have been introduced and used in Korea since 2007 [4]. The vaccines that are licensed and used in Korea are Rotarix (GlaxoSmithKline Biologicals, Rixensart, Belgium) and RotaTeq (Merck Sharp & Dohme Corp., Kenilworth, NJ, USA). Although there are differences in the antigen composition between the 2 vaccines, both are live-attenuated vaccines for oral administration. Live vaccines have a risk of inducing disease, especially among immune-compromised individuals [5]. Many reports have shown that rotavirus vaccination is dangerous, particularly for children with severe combined immunodeficiency (SCID) [6,7]. The introduction of the rotavirus vaccine into the national immunization program (NIP) is under review in Korea, and after its inclusion, the incidence of vaccine-related diseases is expected to increase. Therefore, it is necessary to consider the pathogen characterization process for investigating vaccine-related infection cases. This study reviewed the process of examining a specimen from a SCID patient with confirmed rotavirus infection. In addition, we reviewed the laboratory analysis process to determine whether the vaccine-derived strain of rotavirus has infected a SCID patient.
Materials and Methods
A fecal sample was obtained from a 3-month-old male infant who presented to the Samsung Medical Center, Seoul. He was immunized with RotaTeq at 8 weeks of age and was diagnosed with SCID shortly thereafter. Subsequently, he had persistent diarrhea and was diagnosed with rotavirus infection through a rotavirus test. The patient’s sample was tested at the Korea Disease Control and Prevention Agency to determine whether the rotavirus infection was due to vaccination or another source. The sample was pretreated for a laboratory test. The fecal sample was diluted to 10% (wt/vol) in phosphate-buffered saline (pH 7.2), vortexed for 1 minute, and centrifuged at 8,000 × g for 10 minutes. An antigen test (enzyme immunoassay, EIA) and a genetic test (VP6) were performed on the supernatant fluid after centrifugation to re-confirm the rotavirus infection. RIDASCREEN (R-biopharm AG, Darmstadt, Germany) was used for the EIA test. For the VP6 gene test, nucleic acid was extracted from the fecal sample using a QIAamp Viral RNA Mini Kit (QIAGEN, Hilden, Germany), and reverse-transcription polymerase chain reaction (RT-PCR) was performed [8].
The rotavirus genotype was confirmed using the RT-PCR genotyping method recommended by the World Health Organization (Table 1) [8–10]. The VP4 and VP7 genes were primarily amplified by RT-PCR using nucleic acids extracted from the sample. The second PCR (multiplex PCR) was performed using the primary PCR product as a template. Subsequently, the genotype was determined by confirming the correct amplicon size through an automatic electrophoresis device (Fragment Analyzer Systems; Agilent, Santa Clara, USA) and analysis software (PROSize 3.0; Agilent). Secondary PCR was additionally performed using a single primer for each genotype to investigate the other genotypes unidentified by multiplex PCR. Primary PCR products were also cloned, and the nucleotide sequence was analyzed to confirm whether there was a co-infection involving various genotypes.
The fingerprint gene of RotaTeq, NSP3 (WC3), was detected to determine whether the virus from the patient was the vaccine strain [10]. The nucleotide sequences of VP7 and VP4 from vaccine-derived rotavirus strains were compared with the RotaTeq vaccine strain and human wild-type strains. Multiple sequence alignments were performed with Clustal W, and phylogenetic trees were constructed in MEGA 6.0 using the neighbor-joining tree method, with 1,000 bootstrap replicates based on the Kimura-2 model. The reference sequences used in the phylogenetic analysis were obtained from the GenBank database.
Results
Rotavirus infection was verified, as the sample was positive on EIA and VP6 genetic testing. The genotype was identified as G1P[8] by the RT-PCR genotyping method. Individual PCR testing was also performed for each genotype to identify the other genotypes (G2, G3, G4) present in the same amount in RotaTeq. As a result, G3 was also confirmed. Cloning was performed, but no additional genotypes were identified. Finally, it was confirmed that amplified NSP3 (WC3) was the gene introduced in the vaccine recombination process (Table 2). In addition, the phylogenetic trees constructed for the VP7 and VP4 genes of the RotaTeq vaccine strains (G1, P[8]) revealed that all strains clustered closely together (Figure 1).
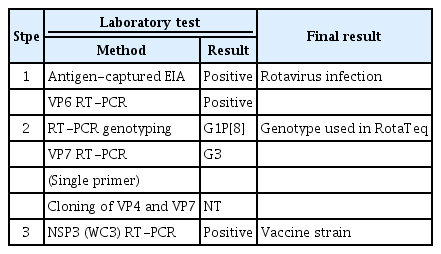
Testing process used for confirming infection due to the rotavirus vaccine strain in an infant with severe combined immunodeficiency
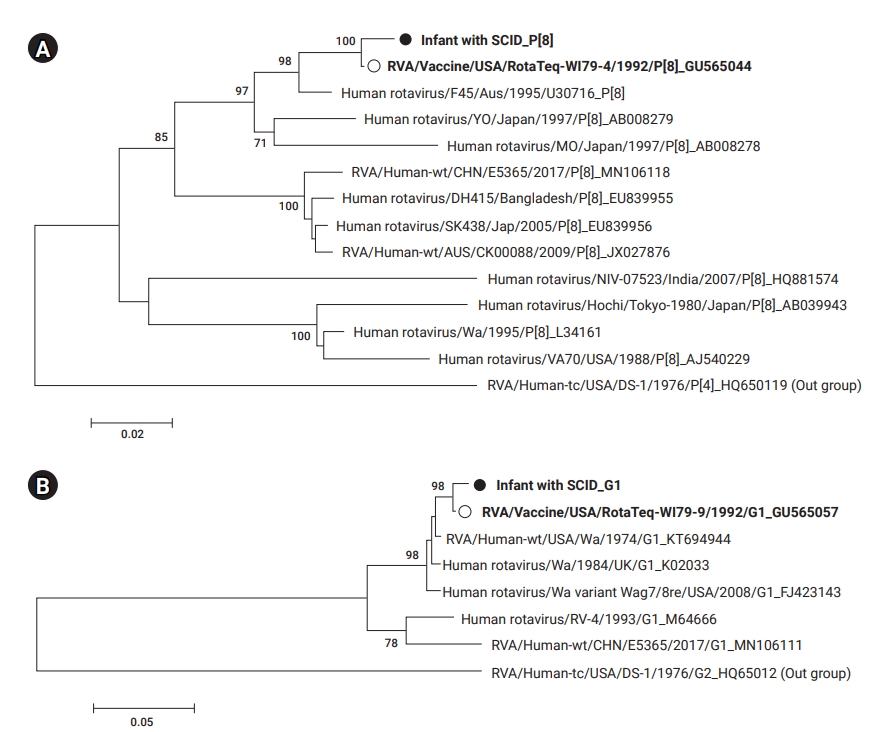
Phylogenetic trees based on the sequences of rotavirus VP4 (A) and VP7 (B) detected from an infant with severe complex immunodeficiency (SCID) (●) compared with (○) RotaTeq (G1 and P[8]). The number at each node indicates the level of bootstrap support (%) based on neighbor-joining analysis of 1,000 resampled datasets. Only values above 70% are displayed.
Discussion
SCID is a genetic condition that results in lack of cellular and humoral immunity. Immunization with rotavirus vaccines (2-dose Rotarix or 3-dose RotaTeq) begins with the first dose, which is administered at 2 months. However, the immune system is usually tested after 4 months of age. Therefore, most SCID infants would have already received a live rotavirus vaccine prior to the diagnosis of SCID. In this case, the patient had been vaccinated with the rotavirus vaccine (RotaTeq) before being diagnosed with SCID and had persistent diarrhea, which is a symptom of rotavirus infection. There have been several reports of rotavirus infection in infants with SCID following vaccination with the RotaTeq live-attenuated vaccine [6,11]. In the United States, the incidence of rotavirus-related diarrhea decreased after the inclusion of rotavirus vaccination in the NIP and then increased again [12]. An analysis of the rotavirus epidemic after the introduction of the NIP confirmed that the vaccine strain was the cause of about 60% of infections [13]. When the rotavirus vaccine is introduced into the NIP, rotavirus-related diarrhea is generally reduced, but infections with the vaccine’s rotavirus strain infection increase to some extent. Children with immune deficiencies such as SCID are at a high risk of vaccine-associated infections.
RotaTeq is a preventive vaccine against rotavirus infection caused by the G1, G2, G3, G4, and P[8] types and comprises 5 recombinant strains (G1P[5], G2P[5], G3P[5], G4P[5], and G6P[8]). Therefore, in this study, tests were conducted to confirm rotavirus infection, genotyping, and identification of the vaccine strains. The rotavirus genotypes detected in the patient were G1 and P[8], which are associated with the vaccine strain. The identification of other genotypes included in equal amounts in the vaccine was attempted, although G1 and P[8] were confirmed using RT-PCR genotyping. It was impossible to identify genotypes other than G1 and P[8] by cloning, but G3 was also confirmed through individual PCR for each genotype. Moreover, it was confirmed that the patient's infection was related to the rotavirus vaccination, as demonstrated by the amplification of the NSP3 (WC3) gene used in the RotaTeq® vaccine for the recombination of vaccine strains.
G1 and P[8] were identified in the genotyping analysis, and rotavirus G1P[8] seemed to be detected. Although G3 was confirmed by PCR using a single primer, it was not possible to confirm the 5 kinds of recombinant strains (G1P[5], G2P[5], G3P[5], G4P[5], and G6P[8]) used in the vaccine. In particular, G1P[8] is the most commonly detected genotype globally, and it is not possible to distinguish whether it is transmitted from vaccines, infected people, or the environment. Some reports have described distinguishing the vaccine strain from the wild-type strain [10,14], and a rapid method using real-time RT-PCR has been recently developed [15]. In this study, the WC3 gene, a fingerprint gene of RotaTeq, was detected, and a thorough analysis of the VP4 and VP7 genes confirmed a close relationship with the vaccine strain. However, the G1P[8] type was not used directly in the RotaTeq vaccine. It was previously found that the patients shed G1P[8] among the 5 surface proteins (G1, G2, G3, G4, P[8]) when infected by the recombinant vaccine [5]. Therefore, based on these findings, the possibility that the detected virus was a wild-type strain could not be confirmed; instead, the detected virus was confirmed to be the vaccine strain.
Conclusion
This study confirmed that the rotavirus infection in SCID patients was related to the vaccine strain by identifying the genotype (G1, G3, P[8]) and detecting the bovine gene (WC3) used in the vaccine strain (RotaTeq). However, it is difficult to identify all the genotypes used in the vaccine immediately, and genetic linkage analysis may be helpful to determine whether there is another wild-strain infection.
Notes
Ethics Approval
Not applicable.
Conflicts of Interest
The authors have no conflicts of interest to declare.
Funding
This study was supported by the Acute Diarrheal Laboratory Surveillance (EnterNet-Korea) in Korea (4800-4851-304). The authors thank the Samsung Medical Center for providing motivation and assistance in the experiment.
Availability of Data
All relevant data are within the manuscript.
Authors’ Contributions
Conceptualization: DYL; Supervision: SJC; Methodology: SRC; Investigation: SRC; Writing–original draft: SJC: Writing–review & editing: WC, MGH; All authors read and approved the final manuscript.