Articles
- Page Path
- HOME > Osong Public Health Res Perspect > Volume 1(1); 2010 > Article
-
Original Article
Molecular Classification of Human Adenovirus Type 7 Isolated From Acute Respiratory Disease Outbreak (ARD) in Korea, 2005–2006 - Wan Ji Lee1, Chun Kang2, Yoon Seok Chung2, Kisoon Kim1
-
Osong Public Health and Research Perspectives 2010;1(1):10-16.
DOI: https://doi.org/10.1016/j.phrp.2010.12.005
Published online: December 7, 2010
1Division of Respiratory Viruses, Center for Infectious Disease, National Institute of Health, Korea Centers for Disease Control and Prevention, Seoul, Korea
2Division of Influenza Virus, Center for Infectious Disease, National Institute of Health, Korea Centers for Disease Control and Prevention, Seoul, Korea
- ∗Corresponding authors. Division of Respiratory Viruses, Center for Infectious Disease, National Institute of Health, Korea Centers for Disease Control and Prevention, 194, Tongil-ro, Eunpyeong-gu, Seoul 122-701, Korea. Division of Influenza Virus, Center for Infectious Disease, National Institute of Health, Korea Centers for Disease Control and Prevention, 194, Tongil-ro, Eunpyeong-gu, Seoul 122-701, Korea. rollingstones@hanmail.nettigerkis@nih.go.kr
- ∗Corresponding authors. Division of Respiratory Viruses, Center for Infectious Disease, National Institute of Health, Korea Centers for Disease Control and Prevention, 194, Tongil-ro, Eunpyeong-gu, Seoul 122-701, Korea. Division of Influenza Virus, Center for Infectious Disease, National Institute of Health, Korea Centers for Disease Control and Prevention, 194, Tongil-ro, Eunpyeong-gu, Seoul 122-701, Korea. rollingstones@hanmail.nettigerkis@nih.go.kr
© 2010 Published by Elsevier B.V. on behalf of Korea Centers for Disease Control and Prevention.
This is an Open Access article distributed under the terms of the Creative Commons Attribution Non-Commercial License (http://creativecommons.org/licenses/by-nc/3.0) which permits unrestricted non-commercial use, distribution, and reproduction in any medium, provided the original work is properly cited.
Abstract
-
Objectives
- To assess the genomic characteristics of human adenoviruses (HAdVs) that caused small-scale epidemics in Korea and compare sequence analysis and restriction fragment length polymorphism (RFLP).
-
Methods
- Two hundred sixty-two throat swabs were collected from geographically distinct two cohabitation facilities during outbreaks in August 2005 and February–May 2006. 148 isolates were obtained using the adenocarcinomic human alveolar basal epithelial cells (A549 cells) from 262 specimens. The sequences of 448 bp partial hexon gene of isolates were analized and compared with serotype results using neutralizing test. The hexon (1.2 kb), fiber, and E4 ORF 6/7 34.7 kDa protein (2.1 kb) genes were further analysed in 10 randomly selected specimens. RFLP of the genomic DNA for genotyping was also performed and compared with sequence information.
-
Results
- All the isolates were localized into the same cluster when phylogenetic tree was generated based on hexon gene using Clustal W. While fiber and E4 ORF 6/7 34.7 kDa protein genes were analysed, the tree was divided into two clusters. Interestingly, isolates with same genetic characteristics of hexon gene did not show identical RFLP patterns in accordance with their origin of episode, rather phylogenetic analysis of fiber and E4 ORF 6/7 34.7 kDa protein genes were correlated with RFLP patterns.
-
Conclusion
- These results indicate that serotype classification based on hexon gene might not be enough to discriminate HAdV serotype, and additional genetic characteristics including fiber and/or E4 ORF 6/7 should be recruited to dispose subgroup of HAdV serotype.
- Human adenoviruses (HAdVs) are approximately 70 nm in diameter and contain a DNA genome. The 51 known HAdV serotypes are geographically ubiquitous, and about one-third of the known HAdV serotypes can cause human diseases with wide range of clinical syndromes. These can be non-respiratory infections involving the stomach, intestine, eyes, and bladder.1 Through nationwide laboratory surveillance system for acute respiratory illness, which has been started since 2005 in Korea, annual average incidence of HAdV accounted for 2.41% of respiratory viruses that cause acute respiratory disease (ARD) during 2006–2009.2 HAdV Types 1, 2, 3, 5, and 7 are often isolated from patients with respiratory tract illness, and ARD is mainly caused by adenovirus infections under circumstance of crowding and stress.3,4 Among major respiratory HAdVs, Type 7 could be further classified into 13 genome types (Types 7a–7l and 7p) that have been distinguished by restriction analysis of genomic DNA.5 Although relationship between genome type and virulence is unclear, the geographical distribution and time of HAdV isolation may be feasible by genome type.3,6,7 HAdV genotyping and serotyping can also be achieved by sequence analysis of the hexon, fiber, and E4 ORF 6/7 34.7 kDa protein gene. The hexon gene consists of conserved and hypervariable regions. Because the hypervariable regions contain serotype-specific residues, sequence analysis of hexon gene allows serotyping of HAdV.8 A previous study documented variation in the fiber and E4 ORF 6/7 34.7 kDa protein genes between two genome types (HAdV Types 7d and 7l) of HAdV Type7 identified as the cause of an outbreak of severe pneumonia in Korea.1
- In this study, we highlighted genetic characteristics of HAdVs that were isolated from two different episodes occurred in Korea, during 2005 and 2006. Genomic diversity was ascertained by sequence analysis of the hexon, fiber, and E4 ORF 6/7 34.7 kDa protein genes, and restriction fragment length polymorphism (RFLP) assay using three restriction enzymes was also performed to compare genome type of isolated HAdVs.
Introduction
- 2.1 Virus culture
- HAdV isolates were obtained by inoculation of clinical specimens onto adenocarcinomic human alveolar basal epithelial cells (A549 cells) cell monolayer in 75 cm2 plastic flasks. When an extensive cytopathic effect was evident (72–96 hours post-infection), cells were treated by three cycles of freezing and thawing. Cell debris was removed by centrifugation at 13,000×g for 5 minutes. Stocks of each virus containing supernatant were kept frozen at −70°C for further analysis. Cells were grown in Dulbecco’s minimum essential medium (Gibco, NY, USA) supplemented with 5% foetal bovine serum (Gibco, NY, USA) and antibiotics and were maintained with Dulbecco’s minimum essential medium supplemented with 2% foetal bovine serum.
- 2.2 Viral DNA extraction
- Supernatant was ultracentrifuged at 20,000×g for 4 hours (SW31 rotor). The virus pellet was suspended in 0.5 mL Tris-EDTA buffer (Welgene, Daegu, Korea) (with 1% sodium dodecyl sulphate (Bio-Rad Laboratories, CA, USA)) and left at 4°C overnight. A 100 μL volume of the suspension was obtained, and 1.0 mL of DNAzol (Molecular Research Center, Inc., OH, USA) was added. Viral DNA was extracted according to the manufacturers’ instructions.
- 2.3 Viral DNA RFLP
- One microgram of full-length viral genomic DNA was used for each of RFLP assay. The DNA was digested with 20U of restriction enzymes BamHI, SmaI, and BstEII (New England Biolabs, MA, USA) for 4 hours, and electrophoresis was carried out using 1% SeaKem Gold agarose (BioWhittaker, ME, USA) in 0.5X Tris-borate-EDTA buffer (Promega,WI, USA) at 2 V/cm for 16 hours with a switching time of 50–90 seconds, using recirculating 0.5X Tris-borate-EDTA buffer. Gels were stained using ethidium bromide.
- 2.4 Polymerase chain reaction and DNA sequencing
- The polymerase chain reaction (PCR) mixture consisted of (1) 1.25U of SP-Taq polymerase; (2) buffer containing 20 mM (NH4)2SO4, 50 mM Tris, and 2.5 mM MgCl2; (3) 0.2 mM of each deoxynucleotide triphosphate mixture (CosmoGenetech, Seoul, Korea), and (4) 10 pmol of each primer (hexon gene; AVUP and AV5L, fiber gene; AV-Fiber-F and AV-Fiber-R, E4 gene; AV-E4-F and AV-E4-R) in a 45 μL reaction volume. The extracted DNA of about 5 μL was added to 45 μL of reaction mixture. PCR cycles consisted of an initial denaturation at 95°C for 3 minutes, followed by 35 cycles of 95°C for 1 minute, 54°C for 1 minute, 72°C for 1 minute, and a final incubation at 72°C for 5 minutes. The products were separated by electrophoresis using 1% agarose gel, stained with SYBR Green I nucleic acid gel stain (Invitrogen, CA, USA) and were observed under ultraviolet light. Amplicons were purified by using QIAquick PCR purification kit (Qiagen, Hilden, Germany) and sequenced in both directions with the BigDye Terminator v3.1 Cycle Sequencing kit (Applied Biosystems, CA, USA). AV3L, AV3R, AVUP, AV5L, AV-Fiber-F, AV-Fiber-R, AV-E4-F, and AV-E4-R were used as sequencing primers (Table 1) for the sequencing reaction as described previously.8 Sequencing products were resolved with an ABI 3730 Genetic analyser (Applied Biosystems, CA, USA). The MEGA 4 program was used for sequence alignment and creating phylogenetic tree, and bootstrap analysis was also performed with neighbour-joining method by resampling of 1000 replicates.
- 2.5 Neutralization test of HAdV
- The neutralization test was performed in 96 well microplates using A549 cells as described previously.9 Briefly, A549 cells were added to wells of a 96-well plate, and a monolayer was allowed to develop. Both 25 μL volumes of HAdV (10 or 100) 50 percents tissue culture infectious dose (TCID50)/mL) and 2-fold dilutions of serum (2.5–0.15U) (Denka Seiken, Tokyo, Japan) were incubated in microtiter plates at 37°C for 90 minutes before addition to the A549 cells. Incubation was continued, and results were recorded until 3–5 days. After 5 days, the 96-well plates were stained with crystal violet solution.
Materials and Methods
- 3.1 Identification of HAdV serotypes by sequence analysis
- To identify the serotype of isolated HAdV, 62 throat swab specimens were collected from one cohabitation facility during an illness outbreak in August 2005 (Episode 1) and 200 specimens were obtained from another facility from February–May 2006 (Episode 2) during another illness outbreak (Table 2). The 448 bp partial region of the hexon gene obtained from 148 isolates (26 isolates in 2005 and 122 isolates in 2006) was amplified and sequenced. Nucleotide sequence analyses revealed that all but one isolate displayed 99% sequence identity with HAdV Type 7. The remained one was HAdV Type 4.
- 3.2 Genetic analysis of the hexon, fiber, and E4 ORF 6/7 34.7-kDa protein genes
- The randomly selected 10 isolates were subjected to analyse genetic characteristics based on genotype and sequences of the hexon (1267 bp), fiber (975 bp), and E4 ORF 6/7 34.7-kDa protein (900 bp) genes. 3.2.1
- There was no variation in the 1267 bp of complete hexon gene among three strains (GQ265844–GQ265846) isolated during the 2005 outbreak and between seven strains (GQ265847–GQ265853) isolated during the 2006 outbreak. The degree of identity with HAdV Type 7 was 90%. Identities of HAdV Types 7a3, 7b, 7c, 7d, 7d2, 7g, 7h, and 7l ranged from 99.3% to 99.9%. The identities of HAdV Types 7d, 7d2, and 7l were 99.9%. However, HAdV Types 7p and 7p1 identities were lower than other HAdV Type 7 (96.1% and 95.9%, respectively) (Figure 1). 3.2.2
- Sequences of the 975-bp fiber gene from the 10 viral isolates were grouped into two clusters with 98.9%–100% identities (Figure 2). The GQ265864 and GQ265866 sequences were identical, whereas other changes were minor (nucleic acid 718 of GQ265865 mutated from A to T, and amino acid 240 mutated from T to S). GQ265864–GQ265866 sequences were similar in sequence to HAdV Type 7l than to HAdV Types 7a, 7d, 7d2, and 7p. The sequences of GQ265867–GQ265873 were identical. In GQ265867, nucleic acid 913 was mutated from C to T, which did not affect the resulting encoded amino acid. GQ265867–GQ265873 sequences closely matched HAdV Types 7d, 7d2, and 7l. Amino acid 112 of GQ265864–GQ265866, GQ265867–GQ265873, and HAdV Type 7l was “V”, on the other hand, HAdV Types 7a, 7d, 7d2, and 7p was “A”. Also, in GQ265867–GQ265873, amino acid 249 was “T”, on the other hand, GQ265864–GQ265866 and HAdV Types 7a, 7d, 7d2, 7p, and 7l was “R”. 3.2.3
- The 900 bp of E4 ORF 6/7 34.7 kDa protein gene was analysed. The GQ265854–GQ265856 sequences and HAdV Type 7l were identical, as was the sequence identity between GQ265857 and GQ265863 sequences and HAdV Types 7d and 7d2. A 98.1% sequence identity was apparent between GQ265854 and GQ265856 and GQ265857 and GQ2658631 (Figure 3). Identical amino acid changes were detected at five sites between GQ265854–GQ265856 and HAdV Type 7d and 7d2 and GQ265857–GQ2658631 and HAdV Type 7l.
- 3.3 Viral DNA RFLP analysis
- DNA of isolated HAdV was digested with restriction enzymes BamHI, SmaI, and BstEII, and electrophoresis was carried out. The resulting BamHI patterns produced two groups: one comprised the isolates in 2005, and the other comprised the isolates in 2006. The BamHI digested RFLP of reference strain HAdV Type 7a did not match neither 2005 nor 2006 isolates. The identical RFLP patterns were obtained by only BstEII digestion among 2005 and 2006 isolates as well as reference strain, whereas distinguished RFLP was obtained by SmaI digestion between reference and isolates (Figure 4).
- 3.4 Neutralization test of HAdV
- The neutralization test was performed using isolated HAdV Type 7 antiserum and HAdV Types 7 and 37. It was found that 100 TCID50 of HAdV Type 7 could infect A549 cells using 0.15–2.5U of antiserum, whereas HAdV Type 37 infection was achieved using 0.3–5U of antiserum. The results demonstrate that the serotype of AF 1–3 and DA 4–10 was HAdV Type7.
Results
3.2.1 Hexon gene
3.2.2 Fiber gene
3.2.3 E4 ORF 6/7 34.7 kDa protein gene
- HAdV is a well-known cause of illness in cohabitation facilities all over the world. To circumvent laborious and time-consuming neutralizing test followed by cell culture for serotyping of the viruses, molecular biological tools such as multiplex PCR, sequencing, and RFLP are widely used. The present study described novel approach to subtype of HAdV Type 7 through RFLP and analysis of viral genes including hexon, fiber, and E4 ORF 6/7 34.7 kDa protein genes.
- Among 262 specimens collected from ARD patients at both episodes, 148 isolates were obtained. Sequence analysis of a portion of the hexon gene determined that 147 strains were HAdV Type 7 and the remaining strain was HAdV Type 4 (data not shown). We randomly selected 10 HAdV Type 7 samples (3 isolates from the 2005 illness outbreak and 7 from the 2006 illness outbreak).
- Serotyping requires sequence analysis of hexon gene because the hypervariable regions of this gene have serotype-specific residues.5,10,11 Confirmatory neutralization test was performed, and 100 and 10 TCID50/mL HAdV Type 37 as a non-matching serotype were capable of infection even in the presence of 5U of anti-HAdV Type 7 serum. On the other hand, 10 TCID50/mL of all the HAdV Type 7 isolates were successfully neutralized in the presence of 0.3U of the antiserum.
- To confirm the genome type of isolated HAdV, the 1267 bp of the hexon gene was amplified and analysed. All 10 of the selected HAdV isolates displayed identical sequences. The 10 isolates did not display identity with other known HAdV Type 7 sequence, but they were much closer (99.9% identity) to HAdV Types 7d, 7d2, and 7l isolates from Korea, Japan, and Israel. More precise genotyping was attempted to sequence analysis of the 975 bp and 900 bp regions of the fiber and E4 ORF 6/7 34.7 kDa protein genes, respectively. The fiber gene-based phylogenetic tree divided the HAdV isolates into two clusters. GQ265864–GQ265866 were genetically most similar to HAdV Type 7l, whereas GQ265868–GQ265873 could not be defined.
- In the phylogenetic tree of the E4 ORF 6/7 34.7 kDa protein genes, more exact genome types were confirmed and also produced two clusters. The sequences of GQ265854–GQ265856 and HAdV Type 7l were identical, as were GQ265857–GQ265863 and Korean isolates HAdV Type 7d and 7d2. E4 ORF 6/7 34.7 kDa protein gene sequences of HAdV 7a and 7p were closer to GQ265857–GQ265863 than were the fiber gene sequences. Through the bootstrap analysis, it was meaningful that the 10 isolates were separated into two clusters by genetic differences.
- RFLP that is used for the general method of classification of genome type of HAdV was also performed using the restriction enzymes BamHI, SmaI, which are classically used in RFLP genome analyses, and BstEII, which allows classification of HAdV Types 7b, 7d2, and 7h.4,6,12–18 The SmaI and BstEII digestions, in common with phylogenetic analysis of hexon gene, produced identical pattern regardless of episodes. In the other hand, BamHI digestion produced two distinctive patterns in accordance with episodes. This BamHI digestive pattern was concomitant with two phylogenetic clusters based on the fiber and E4 ORF 6/7 34.7 kDa protein genes.
- Despite, there might be a self-limitation because we could not perform comparative study with HAdV Types 7d, 7d2, and 7l using the RFLP method. In addition, there might be another possibility of random single-nucleotide mutation, which may have a dramatic effects on restriction patterns, introduced while viruses were being isolated on culture cells. Nevertheless, our results indicate that genetic clusters could be divided differently when different genes were subjected to phylogenetic analysis. Moreover, the dividing clusters of randomly selected isolates were identical to geographical distribution (approximately 160 km) and/or periodical difference of episodes.
- In conclusion, our results presented here indicated that Type 7 HAdV played major causative agent of ARD outbreaks in Korean cohabitation facilities during 2005 and 2006. And the isolates were separated into two HAdV sub-genome Type 7 according to Episodes 1 and 2. With our current knowledge, this is the first report to identify discrepancy of sequence analysis of different genes and RFLP patterns of HAdVs. Simultaneously, present study emphasises that it is of importance to apply multiple methods to classify HAdV serotypes and genome types with purpose of understanding prevalence trend, distribution of genome type of the virus that can cause ARD outbreaks.
Discussion
-
Acknowledgements
- The authors thank the members of the laboratory for their technical support and the patients for their involvement. This study was supported by an intramural research fund (4851-300-210-13) from the Korea Centers for Disease Control and Prevention.
Acknowledgments
-
This is an Open Access article distributed under the terms of the Creative Commons Attribution Non-Commercial License (http://creativecommons.org/licenses/by-nc/3.0) which permits unrestricted non-commercial use, distribution, and reproduction in any medium, provided the original work is properly cited.
Article information
- 1. Cho E.H., Kim H.S., Eun B.W.. Adenovirus type 7 peptide diversity during outbreak, Korea, 1995–2000. Emerg Infect Dis 11:2005;649−654. PMID: 15890114.ArticlePubMedPMC
- 2. Acute infectious agents laboratory surveillance reports (weekly). Available at:. http://www.cdc.go.kr. [Date accessed: October 2010].
- 3. Kim Y.J., Hong J.Y., Lee H.J.. Genome type analysis of adenovirus types 3 and 7 isolated during successive outbreaks of lower respiratory tract infections in children. J Clin Microbiol 41:2003;4594−4599. PMID: 14532188.ArticlePubMedPMC
- 4. Li Q.G., Wadell G.. Analysis of 15 different genome types of adenovirus type 7 isolated in five continents. J Virol 60:1986;331−335. PMID: 3018298.ArticlePubMedPMC
- 5. Crawford-Miksza L.K., Schnurr D.P.. Analysis of 15 adenovirus hexon proteins reveals the location and structure of seven hypervariable regions containing serotype-specific residues. J Virol 70:1996;1836−1844. PMID: 8627708.ArticlePubMedPMC
- 6. Wadell G., Cooney M.K., Linhares A.C.. Molecular epidemiology of adenovirus: global distribution of adenovirus 7 genome types. J Clin Microbiol 21:1985;403−408. PMID: 3980690.ArticlePubMedPMC
- 7. Choi E.H., Lee H.J., Kim S.J.. Ten-year analysis of adenovirus type 7 molecular epidemiology in Korea, 1995–2004: implication of fiber diversity. J Clin Virol 35:2006;388−393. PMID: 16458589.ArticlePubMed
- 8. Crawford-Miksza L.K., Nang R.N., Schnurr D.P.. Strain variation in adenovirus serotypes 4 and 7a causing acute respiratory disease. J Clin Microbiol 37:1999;1107−1112. PMID: 10074533.ArticlePubMedPMC
- 9. Hashido M., Mukouyama A., Sakae K.. Molecular and serological characterization of adenovirus genome type 7h isolated in Japan. Epidemiol Infect 122:1999;281−286. PMID: 10355793.ArticlePubMedPMC
- 10. Casas I., Avellon A., Mosquera M.. Molecular identification of adenoviruses in clinical samples by analyzing a partial hexon genomic region. J Clin Microbiol 43:2005;6176−6182. PMID: 16333124.ArticlePubMedPMC
- 11. Lu X., Erdman D.D.. Molecular typing of human adenoviruses by PCR and sequencing of a partial region of the hexon gene. Arch Virol 151:2006;1587−1602. PMID: 16502282.ArticlePubMed
- 12. Noda M., Yoshida T., Sakaguchi T.. Molecular and epidemiological analyses of human adenovirus type 7 strains isolated from the 1995 nationwide outbreak in Japan. J Clin Microbiol 40:2002;140−145. PMID: 11773107.ArticlePubMedPMC
- 13. Li Q.G., Henningsson A., Juto P.. Use of restriction fragment analysis and sequencing of a serotype-specific region to type adenovirus isolates. J Clin Microbiol 37:1999;844−847. PMID: 9986873.ArticlePubMedPMC
- 14. Ikeda Y., Yamaoka K., Noda M., Ogino T.. Genome types of adenovirus type 7 isolated in Hiroshima city. J Med Virol 69:2003;215−219. PMID: 12683410.ArticlePubMed
- 15. Erdman D.D., Xu W., Gerber S.I.. Molecular epidemiology of adenovirus type 7 in the United States, 1966–2000. Emerg Infect Dis 8:2002;269−277. PMID: 11927024.ArticlePubMedPMC
- 16. Li Q.G., Zheng Q.J., Liu Y.H., Wadell G.. Molecular epidemiology of adenovirus types 3 and 7 isolated from children with pneumonia in Beijing. J Med Virol 49:1996;170−177. PMID: 8818961.ArticlePubMed
- 17. Elnifro E.M., Cooper R.J., Klapper P.E., Bailey A.S.. PCR and restriction endonuclease analysis for rapid identification of human adenovirus subgenera. J Clin Microbiol 38:2000;2055−2061. PMID: 10834953.ArticlePubMedPMC
- 18. Allard A., Albinsson B., Wadell G.. Rapid typing of human adenoviruses by a general PCR combined with restriction endonuclease analysis. J Clin Microbiol 39:2001;498−505. PMID: 11158096.ArticlePubMedPMC
References
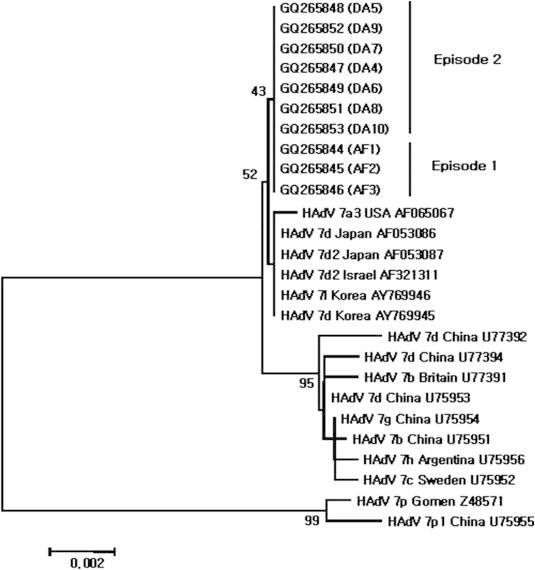
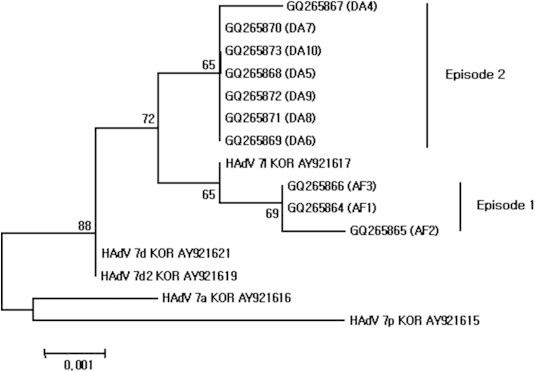

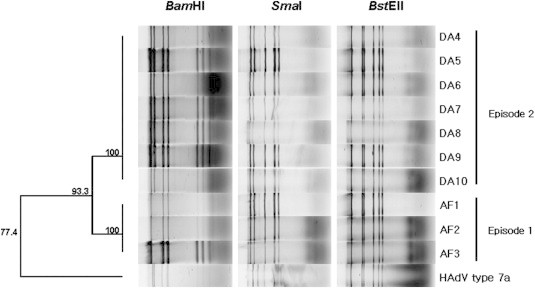
Figure & Data
References
Citations

- Diverse genotypes of human enteric and non-enteric adenoviruses circulating in children hospitalized with acute gastroenteritis in Thailand, from 2018 to 2021
Arpaporn Yodmeeklin, Kattareeya Kumthip, Nuthapong Ukarapol, Hiroshi Ushijima, Niwat Maneekarn, Pattara Khamrin, Day-Yu Chao
Microbiology Spectrum.2023;[Epub] CrossRef - Molecular epidemiology of a post-influenza pandemic outbreak of acute respiratory infections in Korea caused by human adenovirus type 3
Wan-Ji Lee, Hee-Dong Jung, Hyang-Min Cheong, Kisoon Kim
Journal of Medical Virology.2015; 87(1): 10. CrossRef - Antigenic variability among two subtypes of human adenovirus serotype 7
Xingui Tian, Xiaobo Su, Chunyan Xue, Xiao Li, Zhichao Zhou, Rong Zhou
Virus Genes.2014; 49(1): 22. CrossRef


 PubReader
PubReader Cite
Cite
