Articles
- Page Path
- HOME > Osong Public Health Res Perspect > Volume 9(1); 2018 > Article
-
Original Article
Prevalence of Fecal Carriage of CTX-M-15 Beta-Lactamase-ProducingEscherichia coli in Healthy Children from a Rural Andean Village in Venezuela - María Araque, Indira Labrador
-
Osong Public Health and Research Perspectives 2017;9(1):9-15.
DOI: https://doi.org/10.24171/j.phrp.2018.9.1.03
Published online: December 31, 2017
Laboratory of Molecular Microbiology, Department of Microbiology and Parasitology, University of The Andes, Mérida, Venezuela
- *Corresponding author: María Araque, Laboratory of Molecular Microbiology, Department of Microbiology and Parasitology, University of The Andes, Mérida, Venezuela, E-mail: araquemc@ula.ve
• Received: May 31, 2017 • Revised: December 26, 2017 • Accepted: January 8, 2018
Copyright ©2018, Korea Centers for Disease Control and Prevention
This is an open access article under the CC BY-NC-ND license (http://creativecommons.org/licenses/by-nc-nd/4.0/).
Abstract
-
Objectives
- Antimicrobial resistant extended-spectrum-β-lactamase-producing Enterobacteriaceae (ESBL-PE) have been shown to be present in healthy communities. This study examined healthy children from the rural Andean village of Llano del Hato, Mérida, Venezuela, who have had little or no antibiotic exposure to determine the prevalence of fecal carriage of ESBL-producing Escherichia coli (ESBL-EC).
-
Methods
- A total of 78 fecal samples were collected in healthy children aged from 1 to 5 years. ESBL-EC were selected in MacConkey agar plates with cefotaxime and further confirmed by the VITEK 2 system. ESBL were phenotypically detected and presence of bla genes and their variants were confirmed by molecular assays. Determination of phylogenetic groups was performed by PCR amplification. Risk factors associated with fecal carriage of ESBL-EC-positive isolates were analyzed using standard statistical methods.
-
Results
- Of the 78 children studied, 27 (34.6%) carried ESBL-EC. All strains harbored the blaCTX-M-15 allele. Of these, 8 were co-producers of blaTEM-1, blaTEM-5, blaSHV-5 or blaSHV-12. Co-resistance to aminoglycosides and/or fluoroquinolones was observed in 9 strains. 51.9% of ESBL-EC isolates were classified within phylogroup A. A significant, positive correlation was found between age (≥2.5 – ≤5 years), food consumption patterns and ESBL-EC fecal carriage.
-
Conclusion
- This is the first study describing the high prevalence of fecal carriage of ESBL-EC expressing CTX-M-15- among very young, healthy children from a rural Andean village in Venezuela with scarce antibiotic exposure, underlining the importance of this population as a reservoir.
- The spread of extended-spectrum-β-lactamase-producing Enterobacteriaceae (ESBL-PE) is an emerging health problem worldwide [1]. Most genes encoding ESBL are plasmid borne and circulate among bacterial species by horizontal transfer. ESBLs confer resistance to penicillins, cephalosporins, and monobactams, but not to cephamycins or carbapenems and are inhibited by clavulanic acid, tazobactam, and sulbactam [2].
- ESBL-PE have no longer been limited to community-onset or hospital-acquired infections. Fecal carriers of ESBL represent an important reservoir contributing to person-to-person transmission [3]. Antimicrobial resistance in commensal Escherichia coli (E. coli) from healthy communities has been demonstrated for more than 40 years [4]. However, in the last decade high rates of antibiotic-resistant E. coli isolates from healthy children and adults have been reported in different countries. In fact, carriage rates of ESBL-PE in poor children from urban areas in Latin America were as low as 0.1% in 2002, but increased to 1.7% in 2005, and reached 12.4% in 2011 [1,4,5]. A study carried out at 3 urban localities in France revealed an ESBL-PE carrier incidence of 4.6% in asymptomatic children [6]. On the other hand, Bartoloni et al [7] reported that approximately 2/3 of individuals living in a remote Peruvian Amazonas village, with minimal antibiotic exposure, carried resistant E. coli strains to several antibiotics. Hence, fecal E. coli is regarded as a useful indicator of the spread of acquired antibiotic resistance genes in the community [1–4]. In Venezuela, spread of ESBL-PE in hospitals, food-producing healthy animals, and food products have been described [8–13], but little information on the fecal carriage of ESBL-PE in healthy children living in rural or remote communities and with scarce or absent antibiotic exposure is known [14]. The main aim of this study was to investigate the prevalence, phenotypic resistance patterns, and genetic characteristics of commensal ESBL-producing E. coli (ESBL-EC) in healthy children from a rural Andean village in Venezuela.
Introduction
- 1. Ethics statement
- The study was approved by the Committee for Medical and Health Research Ethics of Faculty of Medicine and the Council of Scientific, Humanistic, Technological and Arts (CDCHTA) of the University of The Andes (ULA), Mérida, Venezuela. Written informed consent was obtained from the parents or guardians on behalf of all children before the study started.
- 2. Study design and population
- The study was carried out in the Llano del Hato community, a rural village located in the Municipality of Rangel in the state of Mérida, Venezuela, at 3,538 meters above sea level, at latitude 08° 47,5′ north and longitude 70° 52,4′ west. The population is distributed in 9 rural sectors: El Cabildo, Romeral, Centro, Prado, Lagunita, La Curba, Los Positos, Mesita del Salado and Peña Colorada. The average annual temperature ranges from 2°C to 15°C and has two well-defined climatic seasons: a rainy season between March and October, and a dry season during the rest of the year. The closest permanent health care facility is about 12 km away but due to road conditions, it is usually reached using off-road vehicles, by horse, or by walking. The population is estimated at 438 inhabitants and children under 5 years represent 21.7% (95/438) of that population. The main economic activity is agriculture, with the principal crops being potato, carrots and garlic.
- Between January to July 2015, during a community service study, previously planned by the Faculty of Pharmacy and Bioanalysis of the University of The Andes, 78 children aged from 1 year to 5 years (42 male and 36 female) were selected in a routine clinical check-up with normal findings. The sample size was calculated considering an expected prevalence of 5% and a confidence level of 95% using the Epidat software v3.1 [15]. We excluded children showing symptoms and signs of infection or other diseases, as well as those who received at least one dose of any antibiotic treatment during the previous 15 days. One of each child’s parents signed informed consent and provided demographic information, such as age, sex, crowding (i.e. 2 or more people sleeping in the same room), dietary (dairy products, beef and/or chicken consumption) and hygiene habits. In addition, we asked if there were histories of hospitalizations in the last year, or antibiotic treatments during the previous 3 months.
- 3. Microbiological analysis
- Fecal samples were collected after natural evacuation and an aliquot was immediately inoculated in transport medium and sent to the Molecular Microbiology Laboratory of the University of The Andes at room temperature. Processing started within 6 hours of sample collection. Fecal samples were streaked onto MacConkey agar (Oxoid, Ltd., Basingstoke, UK) with and without cefotaxime (2 mg/L), and aerobically incubated a 37°C for 24 h. When a sample showed positive growth on the selective medium, at least 3 lactose-fermenting pink colonies resembling E. coli were selected for subsequent characterization. E. coli strains were further confirmed by the VITEK 2 Compact system (bioMérieux, Marcy-l’Étoile, France). For the purposes of this work, only one strain of E. coli for each child was selected.
- Antibiotic susceptibility was determined by minimum inhibitory concentration using the VITEK 2 Compact system (AST-GN-299 susceptibility cards) according to the Clinical and Laboratory Standards Institute (CLSI) guidelines [16]. ESBL expression was confirmed by the disc diffusion method on Mueller Hinton agar (Oxoid) using cefotaxime (30 μg) and ceftazidime (30 μg) with and without clavulanic acid (10 μg), as recommended by the CLSI [16]. E. coli ATCC 25922 and Klebsiella pneumoniae ATCC 700603 were used as quality control strains.
- 4. Molecular analysis of ESBL-producing E. coli strains
- DNA was extracted using the X-trem preparation kit (Biotech, Granada, Spain). blaTEM, blaSHV and group blaCTX-M genes were detected by PCR based on previously described primers and protocols [17]. All amplification products were purified (AccuPrep1 PCR Kit; Bioneer, Daejeon, South Korea) and nucleotide sequencing was performed using a 3730XL Genetic Analyzer (Applied Biosystems, Carlsbad, CA). Nucleotide and amino acid sequence alignments were analyzed using the Basic Local Alignment Search Tool (BLAST) suite of programs (https://blast.ncbi.nlm.nih.gov/Blast.cgi). The phylogenetic group of each strain was determined according to Clermont et al [18].
- 5. Statistical analysis
- Data was analyzed using the SPSS version 21 software (IBM Corporation, NY, USA). Categorical data was compared using the Chi-square test with Yates correction. Multivariate logistic regression analysis was used to determine risk factors associated with the fecal carriage of ESBL-EC and results are presented as odd ratios (ORs) with 95% confidence intervals (CIs). Statistical significance was set at p <0.05. Cramer’s V was utilized to determine the effect size or strength of the association between 2 nominal variables and interpretation of the results was carried out as described by Lee [19]. Multivariate analysis of principal components and hierarchical clustering was performed using Past v3.06 program according to Hammer et al [20]. The dendrogram was generated from clinical and epidemiological variable analyses, as well as resistance patterns and genetic characteristics of ESBL-EC isolated in healthy children using the paired Group Algorithm (UPGMA) and Gower similarity index.
Materials and Methods
- 1. Characteristics of the study population and risk factors for fecal carriage
- Of the 78 children included in this study, 42 (53.8%) were males and 36 (46.2%) were females. The median age was 2.4 years (δ=1.1; IQR 1.4–3.3). Table 1 shows the demographic and epidemiological details of children from whom E. coli strains were isolated. Of the 78 fecal samples studied, 27 (34.6%) carried ESBL-EC strains. Of the 44 children aged 1 1 year – ≤ 2.5 years, only 7 were ESBL-EC positive (15.9%), compared with the group aged ≥ 2.5 years – ≤ 5 years, where 20 of 34 children (58.8%) were significantly, associated with the carriage of ESBL-EC (p= 0.0002; OR= 7.551; 95% CI: 2.621 – 21.754). Also, a variety of food consumption was an important factor associated with the risk of carrying ESBL-EC. Children who were breastfed had a lower risk (6 of 37; 16.2%) of carriage of ESBL-EC than those who consumed other food types (21 of 41; 51.2%) (p= 0.0026; OR= 5.425; 95% CI: 1.865 – 15.778). Five out of 6 children who received antibiotics within the last 3 months had a higher risk of ESBL-EC carriage (p= 0.0304; OR= 11.364; 95% CI: 1.252–103.11).The Cramer’s V values confirmed a relatively strong effect size (0.447) of the variable ‘≥ 2.5 – ≤5 years,’ and a moderate effect (0.367) of the variable ‘food consumption variety’ as favorable conditions for carrying ESBL-EC. On the other hand, Cramer’s V values for antibiotic consumption (0.296) or hospital admission (0.124) had a low effect (0.296) on the fecal carriage of ESBL-EC in the study population (Figure 1).
- 2. Antibiotic susceptibility phenotype
- Antimicrobial susceptibility of 78 E. coli strains isolated from healthy children from the Llano del Hato community is shown in Table 2. Antibiotic-resistant E. coli were detected in 34 (43.6%) of the 78 strains studied. Resistance to wide-spectrum cephalosporins was observed in more than 33% of strains, while resistance to fluoroquinolones and aminoglycosides spanned from 5.1% to 7.7%. The intermediate susceptibility phenotype was observed only with amikacin, nalidixic acid and ciprofloxacin. All strains were susceptible to piperacillin/tazobactam, carbapenems, tigecycline and colistin.
- Phenotypic assays to screen potential-ESBL producers revealed 27 (34.6%) strains with ESBL activity. Of these, 17 (63%) were exclusive producers of ESBL, and 10 ESBL-EC strains showed co-resistance to, 1 or 2 antimicrobial classes (aminoglycosides and fluoroquinolones). All intermediate susceptibility phenotypes were observed in non-ESBL-EC. Single resistance to nalidixic acid was only detected in one non-ESBL-EC strain (Table 3).
- 3. ESBL genotypes and phylogenetic groups of ESBL-EC
- Molecular characterization of ESBL-EC strains revealed the presence of at least 3 bla genes. All 27 ESBL-EC strains harbored the blaCTX-M-15 allele. Of these, 8 strains co-produced other β-lactamases, 3 (11.1%) co-produced TEM-1b, 2 (7.4% respectively) co-produced TEM-5 and SHV-5, and 1 (3.7%) co-produced SHV-12. Nineteen (70.4%) strains only produced CTX-M-15. Most ESBL-EC strains (14/27; 51.9%) were classified within phylogenetic group A, 8 (29.6%) were members of group B2, 3 (11.1%) of group B1, and 2 (7.4%) of group D (Table 4).
- Fecal carriage distribution of ESBL-EC isolates, according to global relationships of clinical-epidemiological variables, as well as phenotypic and genetic characteristics of E. coli strains are shown in Figure 2. Children were divided into 2 principal clusters with a similarity relationship of approximately 60%. The major cluster was composed of 21 (77.8%) children and the minor cluster by 6 (22.2%) children. Regardless of the epidemiological variables studied, no particular population distribution was observed in relation to the phenotypic or molecular characteristics of E. coli isolates, except 2 subgroups of 4 and 2 children, respectively, located in the major cluster, which presented very similar epidemiological characteristics (100%) and also had CTX-M-15-producing E. coli from the phylogenetic group A as a common characteristic.
Results
- Although E. coli isolates producing ESBLs have previously been reported in healthy people, the majority of subjects analyzed, lived in cities [1,3]. At present, there is little information about isolation rates of ESBL-PE in healthy children from rural or remote communities with scarce or absent antibiotic exposure, as well as risk factors associated with fecal carriage [3,7,21]. To our knowledge, this is the first study describing the prevalence of ESBL-EC fecal carriage among very young, healthy children from a rural Andean village in Venezuela. Results showed a high prevalence (34.6%) of fecal carriage by ESBL-EC in 1 year to 5-year-old healthy children from the Llano del Hato community, which is comparable to the prevalence of 34.3% found among community and hospitalized children in Dar es Salaam, Tanzania [22], worse than values reported in healthy children from Gipuzkoa, Spain (24.0%) [23], and almost 3 times higher than those reported in healthy children in resource-limited settings from Latin American (12%) [24], but lower than prevalence described in asymptomatic rural children in Taian, China (73.9%) [2].
- The rapid spread of the genotype CTX-M-15 has been the subject of worldwide concern in the microbiology community [1,5]. In this study, molecular characterization of CTX-M-type ESBL revealed that 27 strains of ESBL-EC carried blaCTX-M-15. Of these, 8 were co-producers of other ESBLs such as blaTEM-1, blaTEM-5, blaSHV-5 or blaSHV-12. The high prevalence of the CTX-M-15-like genotype further confirms that the CTX-Ms is the dominant enzyme among carriers, both in Venezuela and worldwide [1,11,25]. The strains were distributed in the main phylogenetic groups A, B1, B2 and D, but more than half belonged to phylogroup A. Previous studies performed in Spain [23], France [6] and India [26] reported a higher frequency of phylogroup A in strains of CTX-M-producing E. coli isolated from asymptomatic carriers. Specific association between phylogenetic groups and variants of CTX-M have been suggested, particularly bla genes encoding CTX-M-14 and CTX-M-15 which were considered to be driven by epidemic E. coli strains belonging to phylogroup B2 [27]. However, of the 27 CTX-M-15-producing E. coli strains in this study, only 8 isolates belonged to phylogroup B2. ESBL-producing isolates, especially CTX-M-producing E. coli, exhibit an alarming trend where there is an increasing number of E. coli strains with co-resistance to other classes of antibiotics [1,5]. In this study, the resistance to aminoglycosides and fluoroquinolones in ESBL-producing strains was higher than in non- ESBL-producers. However, the number of fluoroquinolone-resistant strains may be greater if molecular tests had been used for screening. Clinical isolates harboring plasmid-mediated quinolone resistance (PMQR) determinants did not show any significant change in their minimum inhibitory concentration when compared with isolates susceptible to nalidixic acid [13]. Hence, it is necessary to increase test sensitivity and optimize the screening procedures when strains that might contain such resistance determinants are studied. In this regard, it is important to note that in recent decades, an increase in resistance to fluoroquinolones associated with expanded-spectrum cephalosporins in E. coli has been reported in Latin America [5]. Multidrug resistance profiles involving non-β-lactam antibiotics in ESBL-producing isolates may also contribute to the increase in colonization pressure [28]. Considering the origin of the strains studied here, we believe that the source of resistance to fluoroquinolones should not be directly related to the use of quinolones, since these drugs are not prescribed in pediatric patients. However, fluoroquinolones are widely used in veterinary medicine and poultry production, and fluoroquinolone-resistant strains could indeed be transmitted to humans through the food chain [3,13,28].
- Several studies have reported that age is a risk factor for ESBL carriage, but the impact of this epidemiological variable on ESBL fecal carriage varies within the age range of the studied population and also the study settings [22]. Although this work included young children with a limited age range of ≥2.5 years to ≤5 years, this group was significantly associated with ESBLs carriage, and 7 times more likely to be colonized with ESBL-EC than the 1 year to ≤2.5 year group. This finding bears a direct relation to the fact that older children have more varied eating habits, which include different food types. There is considerable evidence demonstrating the role of the food chain in the colonization and spread of ESBL-EC in community settings [4,13,28]. Multivariate analysis revealed a positive association between food consumption patterns and high prevalence of ESBL-EC fecal carriage in healthy children. According to the data recorded in this study, foods regularly consumed by these children included dairy products and meats, mostly home-made or acquired by exchange with nearby villages. Although CTX-M-15 has spread pandemically in humans, this enzyme has recently been reported in food-producing animals in South America [29]. In this regard, previous studies examining the presence of ESBL-PE in dairy products and meats marketed in Venezuela have determined the impact of food products both as reservoirs, and dissemination mechanisms of these strains [12,13,30].
- Unlike other studies [31], we showed that before hospitalization or previous use of antimicrobial drugs, there were risk factors for fecal carriage of ESBL-EC in healthy children that had little impact. Also, it was not evident that other variables such as gender, hand washing and household crowding played a major role in the individual carriage of ESBL-EC. These results highlight that additional factors influence dissemination of ESBL-EC in healthy children from the Llano del Hato community and emphasizes how limited resistance control strategies are based exclusively on antibiotic restriction policies. Clemente et al. [32] pointed out that functional resistance genes probably are a feature of the human microbiome, even in the absence of exposure to commercial antibiotics. Moreover, resistance genes are primed for dissemination upon exposure to pharmacological levels of antibiotics. Results in this study seem to fit these hypotheses because ESBLs, and particularly CTX-M genes, could be introduced into the community and then disseminated among the population by cross-transmission, as it has been suggested in other studies [7,21]. In this respect, it is important to point out that resistance patterns observed in the Llano del Hato community were globally comparable to those reported in previous studies conducted in urban areas of Mérida State [10,11,25].
- In general, the majority of children with ESBL-EC fecal carriage constituted a heterogeneous population according to epidemiological variable analysis. The genetic characteristics of ESBL-EC strains did not determine a particular distribution in the studied population. Only 6 children with fecal carriage of CTX-M-15-producing E. coli (phylogroup A) shared highly similar characteristics. This is probably because these children were members of close family groups. These results indicate that the epidemiology of colonization with ESBL-producing E. coli is complex and that explaining the dynamics of acquisition and transmission requires complex longitudinal studies.
- Our study had several limitations. We did not determine the presence of plasmids encoding the ESBLs, which could possibly have improved the understanding of the dynamics of ESBL propagation, nor did we determine the presence of virulence factors in the isolates. These studies will be performed shortly. Conversely, the degree of person-to-person contact between relatives was not investigated in detail and, consequently we were unable to assess whether the different types of contact were associated with transmission risks.
- In conclusion, the prevalence of ESBL-EC fecal carriage among healthy children from Llano del Hato is alarmingly high, underlining the importance of this population as a reservoir. CTX-M-15 was the predominant ESBL, which coexisted in some strains with other ESBLs (blaTEM-1, blaTEM-5, blaSHV-5 or blaSHV-12.) A significant, positive correlation was found between age, food consumption patterns and ESBL-EC fecal carriage. Local, national and worldwide research is crucial, since ESBL-EC fecal carriage contributes to a considerable circulating pool of resistance genes. The presence of ESBL-EC fecal carriage in the Llano del Hato community might thus provide valuable information for understanding dissemination in rural villages with low levels of antibiotic exposure.
Discussion
-
Acknowledgements
- This study was partially supported by the Council of Scientific, Humanistic, Technological and Arts of University of The Andes (CDCHTA-ULA), Mérida, Venezuela (grant CVI-ADG-FA-02-97) and Fundación Empresas Polar, Caracas, Venezuela (Project N° 140275).
Acknowledgements
-
Conflicts of Interest
No potential conflict of interest relevant to this article was reported.
Article information
- 1. Woerther PL, Burdet Ch, Chachaty E, Andremont A. Trends in human fecal carriage of extended-spectrum-lactamases in the community: toward the globalization of CTX-M. Clin Microbiol Rev 2013;26(4). 744−58. PMID: 10.1128/CMR.00023-13. PMID: 24092853. PMID: 3811232.ArticlePubMedPMC
- 2. Zhang H, Zhou Y, Guo S, Chang C. High prevalence and risk factors of fecal carriage of CTX-M type extended-spectrum beta-lactamase-producing Enterobacteriaceae from healthy rural residents of Taian, China. Front Microbiol 2015;6(239). 1−5.ArticlePubMedPMC
- 3. Hijazi SM, Fawzi MA, Ali FM, Abd El Galil KH. Prevalence and characterization of extended-spectrum beta-lactamases producing Enterobacteriaceae in healthy children and associated risk factors. Ann Clin Microbiol Antimicrob 2016;15(3). 1−9. PMID: 10.1186/s12941-016-0121-9.ArticlePubMedPMC
- 4. Rolain JM. Food and human gut as reservoirs of transferable antibiotic resistance encoding genes. Front Microbiol 2013;4:173PMID: 10.3389/fmicb.2013.00173. PMID: 23805136. PMID: 3690338.ArticlePubMedPMC
- 5. Bartoloni A, Pallecchi L, Riccobono E, et al. Relentless increase of resistance to fluoroquinolones and expanded-spectrum cephalosporins in Escherichia coli: 20 years of surveillance in resource-limited settings from Latin America. Clin Microbiol Infect 2013;9(4). 356−61. PMID: 10.1111/j.1469-0691.2012.03807.x.Article
- 6. Birgy A, Cohen R, Levy C, Bidet P, Courroux C, Benani M, Thollot F, Bingen E. Community faecal carriage of extended spectrum beta-lactamase-producing Enterobacteriaceae in French children. BMC Infect Dis 2012;12(315). 1−5. PMID: 10.1186/1471-2334-12-315.ArticlePubMedPMC
- 7. Bartoloni A, Pallecchi L, Rodríguez H, et al. Antibiotic resistance in a very remote Amazonas community. Int J Antimicrob Agents 2009;33:125−9. PMID: 10.1016/j.ijantimicag.2008.07.029.ArticlePubMed
- 8. Araque M, Nieves B, Lauretti L, Rossolini GM. Molecular basis of extended-spectrum b-lactamase production in nosocomial isolates of Klebsiella pneumoniae from Mérida, Venezuela. Int J Antimicrob Agents 2000;15(1). 37−42. PMID: 10.1016/S0924-8579(99)00168-5. PMID: 10856675.ArticlePubMed
- 9. Millán B, Castro D, Araque M, Ghiglione B, Gutkind G. ISCR1 asociado a genes blaCTX-M-1 y blaCTX-M-2 en plásmidos IncN e IncFIIA aislados en Klebsiella pneumoniae de origen nosocomial en Mérida, Venezuela. Biomédica 2013;33(2). 268−75.
- 10. Abreu S, Varela Y, Millán B, Araque M. Klebsiella pneumoniae y Escherichia coli productoras de beta-lactamasas de espectro extenso aisladas en pacientes con infección asociada a los cuidados de la salud en un hospital universitario. Enf Inf Microbiol 2014;34(3). 92−9.
- 11. Hernández E, Araque M, Millán Y, Millán B, Vielma S. Prevalencia de β-lactamasa CTX-M-15 en grupos filogenéticos de Escherichia coli uropatógena aisladas en pacientes de la comunidad de Mérida, Venezuela. Invest Clin 2014;55(1). 32−43. PMID: 24758100.PubMed
- 12. Guillén L, Millán B, Araque M. Caracterización molecular de cepas de Escherichia coli aisladas de productos lácteos artesanales elaborados en Mérida, Venezuela. Infectio 2014;18:100−8. PMID: 10.1016/j.infect.2014.04.004.Article
- 13. González F, Araque M. Association of transferable quinolone resistance determinants qnrB19 with extended-spectrum β-lactamases in Salmonella Give and Salmonella Heidelberg in Venezuela. Int J Microbiol 2013;628185.
- 14. Velasco J, González F, Díaz T, Peña-Guillén J, Araque M. Profiles of enteropathogens in asymptomatic children from indigenous communities of Mérida, Venezuela. J Infect Dev Ctries 2011;5(4). 278−85. PMID: 21537069.ArticlePubMed
- 15. Epidat 3.1: Análisis epidemiológico de datos tabulados. A Coruña. Washington, DC: Xunta de Galicia. Organización Panamericana de la Salud (OPS-OMS); 2006. p 20.
- 16. Clinical and Laboratory Standards Institute. CLSI. Performance standard for antimicrobial disk susceptibility testing. 26rd ed. CLSI; supplement M100S. Wayne, PA: 2016.
- 17. Flores-Carrero A, Labrador I, Paniz-Mondolfi A, Peaper DR, Towle D, Araque M. Nosocomial outbreak of ESBL-producing Enterobacter ludwigii coharboring CTX-M-8, SHV-12 and TEM-15 in a neonatal intensive care unit in Venezuela. J Glob Antimicrobial Resist 2016;7:114−8. PMID: 10.1016/j.jgar.2016.08.006.Article
- 18. Clermont O, Bonacorsi S, Bingen E. Rapid and simple determination of the Escherichia coli phylogenetic group. Appl Environ Microbiol 2000;66(10). 4555−8. PMID: 10.1128/AEM.66.10.4555-4558.2000. PMID: 11010916. PMID: 92342.ArticlePubMedPMC
- 19. Lee DY. Alternatives to P value: confidence interval and effect size. Korean J Anesthesiol 2016;69(6). 555−62. PMID: 10.4097/kjae.2016.69.6.555. PMID: 27924194. PMID: 5133225.ArticlePubMedPMCPDF
- 20. Hammer Ø, Harper DAT, Ryan PD. Past: Paleontological statistics software package for education and data analysis. Palaeontologia Electronica 2001 4(1). http://palaeo-electronica.org/2001_1/past/issue1_01.htm.
- 21. Ruppé E, Woerther PL, Diop A, et al. Carriage of CTX-M-15-producing Escherichia coli isolates among children living in a remote village in Senegal. Antimicrob Agents Chemother 2009;53(7). 3135−7. PMID: 10.1128/AAC.00139-09. PMID: 19364858. PMID: 2704652.ArticlePubMedPMC
- 22. Tellevik MG, Blomberg B, Kommedal Ø, et al. High prevalence of faecal carriage of ESBL-producing Enterobacteriaceae among children in Dar es Salaam, Tanzania. PLoS One 2016;11(12). e0168024PMID: 10.1371/journal.pone.0168024. PMID: 27936054. PMID: 5148075.ArticlePubMedPMC
- 23. Fernández-Reyes M, Vicente D, Gomariz M, Esnal Olatz, Landa Joseba, Oñate Eider, Pérez-Trallero Emilio. High rate of fecal carriage of extended-spectrum-β-lactamase-producing Escherichia coli in healthy children in Gipuzkoa, northern Spain. Antimicrob Agents Chemother 2014;58(3). 1822−4. PMID: 10.1128/AAC.01503-13. PMID: 3957886.ArticlePubMedPMC
- 24. Pallecchi L, Bartoloni A, Fiorelli C, et al. Rapid dissemination and diversity of CTX-M extended-spectrum beta-lactamase genes in commensal Escherichia coli isolates from healthy children from low resource settings in Latin America. Antimicrob Agents Chemother 2007;51(8). 2720−5. PMID: 10.1128/AAC.00026-07. PMID: 17548490. PMID: 1932529.ArticlePubMedPMC
- 25. Millán Y, Hernández E, Millán B, Araque M. Distribución de grupos filogenéticos y factores de virulencia en cepas de Escherichia coli uropatógena productora de beta-lactamasa CTX-M-15 aisladas de pacientes de la comunidad en Mérida, Venezuela. Rev Argent Microbiol 2014;46(3). 175−81. PMID: 25444124.PubMed
- 26. Dureja C, Mahajan S, Raychaudhuri S. Phylogenetic distribution and prevalence of genes encoding class I integrons and CTX-M-15 extended-spectrum β-lactamases in Escherichia coli isolates from healthy humans in Chandigarh, India. PLoS One 2014;9(11). e112551PMID: 10.1371/journal.pone.0112551. PMID: 4237346.ArticlePubMedPMC
- 27. Dubois V, De Barbeyrac B, Rogues AM, et al. CTX-M-producing Escherichia coli in a maternity ward: a likely community importation and evidence of mother-to-neonate transmission. J Antimicrob Chemother 2010;65(7). 1368−71. PMID: 10.1093/jac/dkq153. PMID: 20466850.ArticlePubMedPDF
- 28. Bartoloni A, Pallecchi L, Riccobono E, et al. Relentless increase of resistance to fluoroquinolones and expanded spectrum cephalosporins in Escherichia coli: 20 years of surveillance in resource-limited settings from Latin America. Clin Microbiol Infect 2012;19(4). 356−61. PMID: 10.1111/j.1469-0691.2012.03807.x. PMID: 22414066.ArticlePubMed
- 29. Silva KC, Moreno M, Cabrera C, Spira B, Cerdeira L, Lincopan N, Moreno AM. First characterization of CTX-M-15-producing Escherichia coli strains belonging to sequence type (ST) 410, ST224, and ST1284 from commercial swine in South America. Antimicrob Agents Chemother 2016;60(4). 2505−8. PMID: 10.1128/AAC.02788-15. PMID: 26824955. PMID: 4808232.ArticlePubMedPMC
- 30. Molina N, Millán B, Araque M. Indicadores de calidad sanitaria y fenotipificación de Salmonella enterica aislada en pollo crudo comercializado en supermercados del área urbana del estado Mérida, Venezuela. Infectio 2010;14:174−85. PMID: 10.1016/S0123-9392(10)70109-0.Article
- 31. Bryce A, Costelloe C, Hawcroft C, Wootton M, Hay AD. Faecal carriage of antibiotic resistant Escherichia coli in asymptomatic children and associations with primary care antibiotic prescribing: a systematic review and meta-analysis. BMC Infect Dis 2016;16:359PMID: 10.1186/s12879-016-1697-6. PMID: 27456093. PMID: 4960702.ArticlePubMedPMCPDF
- 32. Clemente JC, Pehrsson EC, Blaser MJ, et al. The microbiome of uncontacted Amerindians. Sci Adv 2015;1(3). e1500183PMID: 10.1126/sciadv.1500183. PMID: 26229982. PMID: 4517851.ArticlePubMedPMC
References
Figure 1
Forest plot showing the association of the fecal carriage of ESBL-EC with epidemiological variables.
The values of Cramer’s V confirmed a relatively strong effect size (0.447) of the variable ≥ 2.5 years – ≤5 years, and a moderate effect (0.367) of the variable “food ingestion of animal origin.”
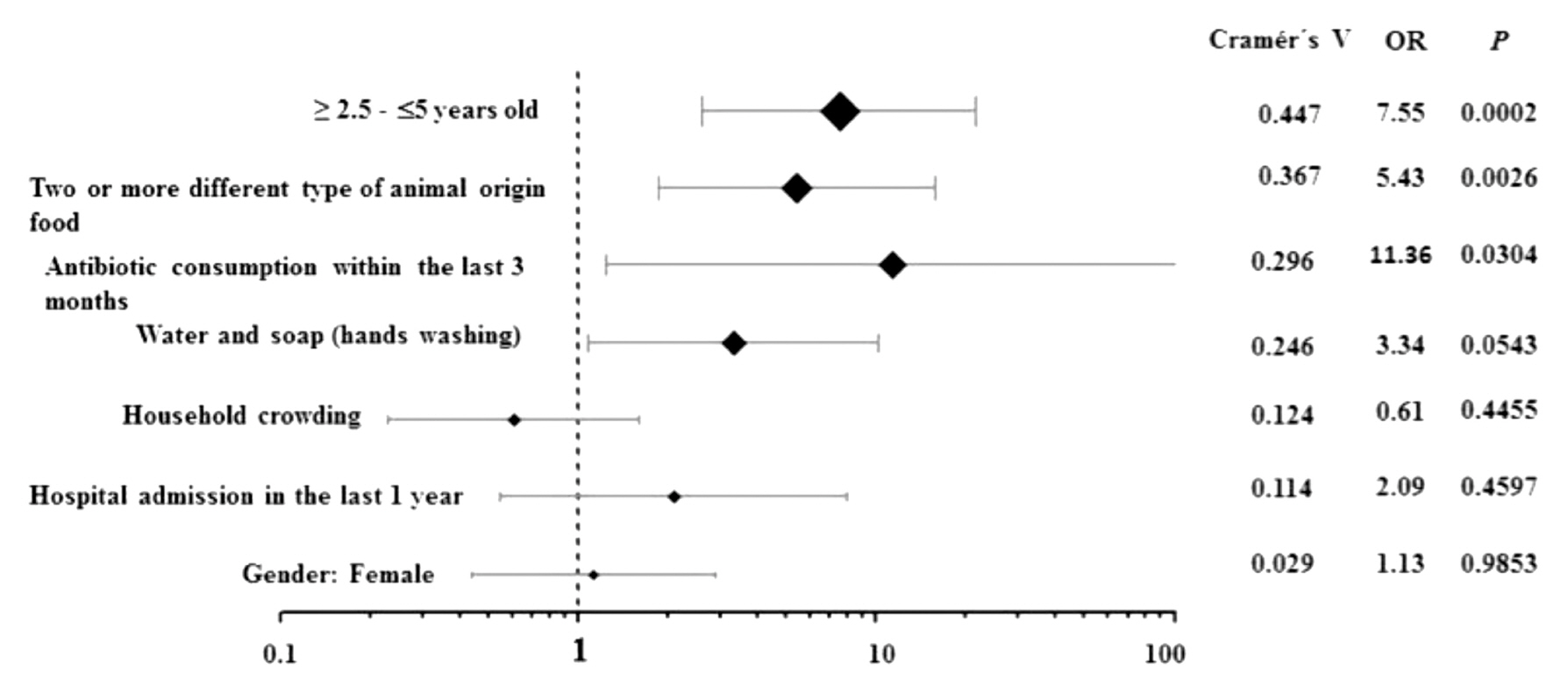

Figure 2
Hierarchical clustering dendogram demonstrating phylogenetic and resistance profiling of ESBL-EC.
Analysis based on clinical, epidemiological and molecular characteristics identified 2 distinct clusters of child fecal carriage of ESBL-EC. Note the presence of 2 subgroups (major cluster) with similar relationships of 100%. Children with a lower degree of similarity (≤65%) were observed in the minor cluster.
AMK: amikacin; CIP: ciprofloxacin; GEM: gentamicin; N°: children (ESBL-EC positive); NAL: nalidixic acid; PG: phylogenetic group; Rest. Profile: resistance profile.
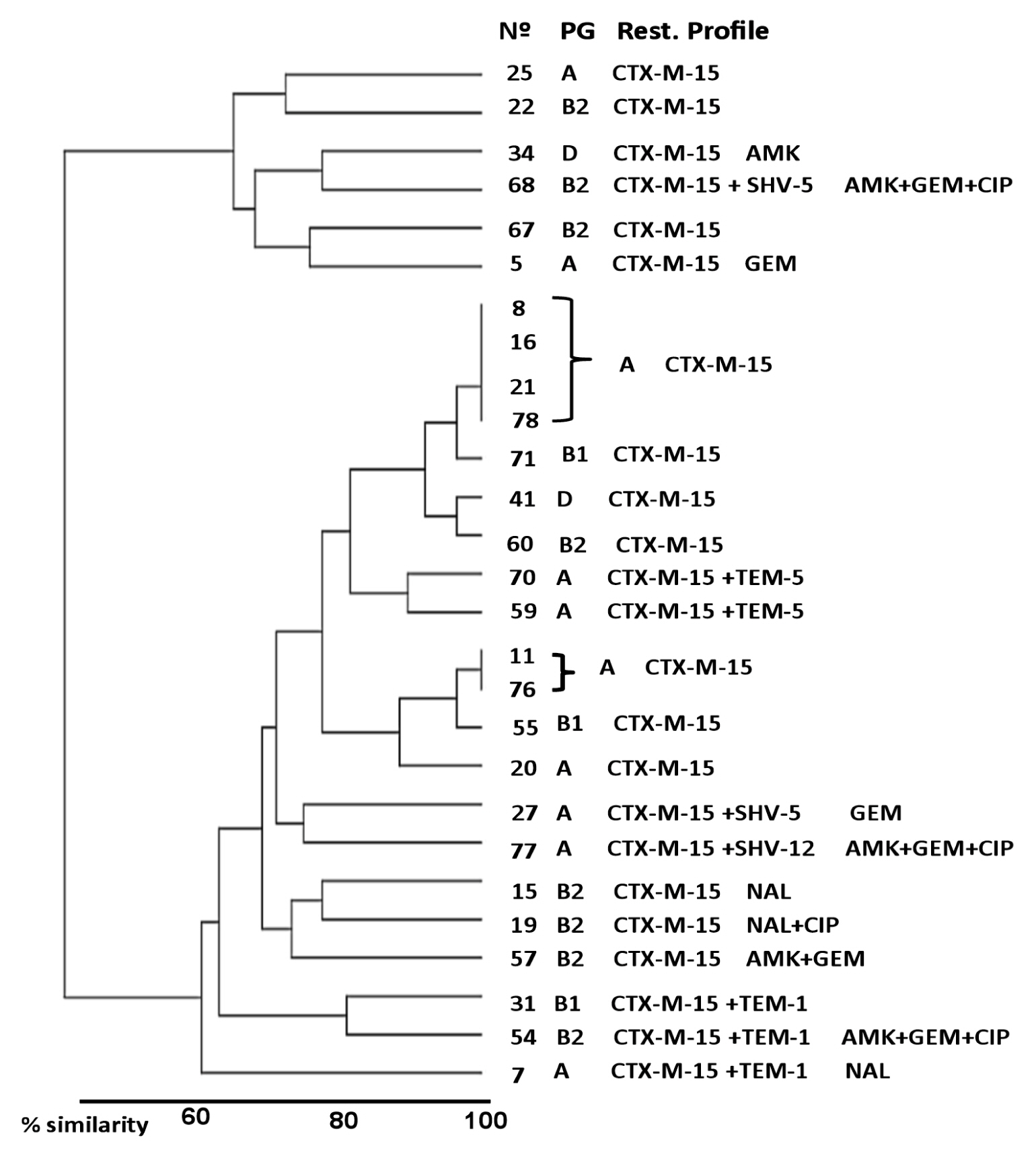

Table 1Demographic and epidemiological characteristics associated with ESBL-producing E. coli from healthy children from a rural Andean village in Venezuela.
Table 2Antimicrobial susceptibility of E. coli strains isolated from healthy children from a rural Andean village in Venezuela.
Table 3Susceptibility profile of E. coli strains isolated from healthy children from a rural Andean village in Venezuela.
Table 4Distribution of blaESBLs genes according to the phylogenetic group of E. coli strains.
Figure & Data
References
Citations
Citations to this article as recorded by 

- First Report of IMI-2-Producing Enterobacter bugandensis and CTX-M-55-Producing Escherichia coli isolated from Healthy Volunteers in Tunisia
Rym Ben Sallem, Ameni Arfaoui, Afef Najjari, Isabel Carvalho, Abdelmalek Lekired, Hadda-Imen Ouzari, Karim Ben Slama, Alex Wong, Carmen Torres, Naouel Klibi
Antibiotics.2023; 12(1): 116. CrossRef - Co-Harboring of Beta-Lactamases and mcr-1 Genes in Escherichia coli and Klebsiella pneumoniae from Healthy Carriers and Backyard Animals in Rural Communities in Ecuador
Carlos Bastidas-Caldes, Emily Cisneros-Vásquez, Antonella Zambrano, Andrea Mosquera-Maza, William Calero-Cáceres, Joaquín Rey, Yoshimasa Yamamoto, Mayumi Yamamoto, Manuel Calvopiña, Jacobus H. de Waard
Antibiotics.2023; 12(5): 856. CrossRef - Extended-Spectrum Beta-Lactamase- and Carbapenemase-Producing Enterobacteriaceae Family of Bacteria from Diarrheal Stool Samples in Northwest Ethiopia
Minichil Worku, Michael Getie, Feleke Moges, Alem Getaneh Mehari, Jayaraman Tharmalingam
Interdisciplinary Perspectives on Infectious Disea.2022; 2022: 1. CrossRef - GENOMIC CHARACTERIZATION OF MULTIDRUG-RESISTANT EXTENDED-SPECTRUM β-LACTAMASE–PRODUCING ESCHERICHIA COLI AND KLEBSIELLA PNEUMONIAE FROM CHIMPANZEES (PAN TROGLODYTES) FROM WILD AND SANCTUARY LOCATIONS IN UGANDA
Sandra L. Bager, Isaac Kakaala, Egle Kudirkiene, Denis K. Byarugaba, John Elmerdahl Olsen
Journal of Wildlife Diseases.2022;[Epub] CrossRef - Relevant increase of CTX-M-producing Escherichia coli carriage in school-aged children from rural areas of the Bolivian Chaco in a three-year period
Selene Rebecca Boncompagni, Maria Micieli, Tiziana Di Maggio, Antonia Mantella, Anna Liz Villagrán, Tatiana Briggesth Miranda, Carmen Revollo, Veronica Poma, Herlan Gamboa, Michele Spinicci, Marianne Strohmeyer, Alessandro Bartoloni, Gian Maria Rossolini,
International Journal of Infectious Diseases.2022; 121: 126. CrossRef - Comparison of the global prevalence and trend of human intestinal carriage of ESBL-producing Escherichia coli between healthcare and community settings: a systematic review and meta-analysis
Yihienew M. Bezabih, Alemayehu Bezabih, Michel Dion, Eric Batard, Samson Teka, Abiy Obole, Noah Dessalegn, Alelegn Enyew, Anna Roujeinikova, Endalkachew Alamneh, Corinne Mirkazemi, Gregory M. Peterson, Woldesellassie M. Bezabhe
JAC-Antimicrobial Resistance.2022;[Epub] CrossRef - Fecal Carriage of Extended-Spectrum β-Lactamases and pAmpC Producing Enterobacterales in an Iranian Community: Prevalence, Risk Factors, Molecular Epidemiology, and Antibiotic Resistance
Nasrin Habibzadeh, Hadi Peeri Doghaheh, Meysam Manouchehri Far, Hossein Alimohammadi Asl, Sohrab Iranpour, Mohsen Arzanlou
Microbial Drug Resistance.2022; 28(9): 921. CrossRef - Extended-Spectrum Beta-Lactamases Producing Escherichia coli in South America: A Systematic Review with a One Health Perspective
Carlos Bastidas-Caldes, Daniel Romero-Alvarez, Victor Valdez-Vélez, Roberto D Morales, Andrés Montalvo-Hernández, Cicero Gomes-Dias, Manuel Calvopiña
Infection and Drug Resistance.2022; Volume 15: 5759. CrossRef - The global prevalence and trend of human intestinal carriage of ESBL-producing Escherichia coli in the community
Yihienew M Bezabih, Wilber Sabiiti, Endalkachew Alamneh, Alamneh Bezabih, Gregory M Peterson, Woldesellassie M Bezabhe, Anna Roujeinikova
Journal of Antimicrobial Chemotherapy.2021; 76(1): 22. CrossRef - Global prevalence and molecular characterization of extended-spectrum β-lactamase producing-Escherichia coli in dogs and cats – A scoping review and meta-analysis
Marília Salgado-Caxito, Julio A. Benavides, Aiko D. Adell, Antonio Carlos Paes, Andrea I. Moreno-Switt
One Health.2021; 12: 100236. CrossRef - Can breastfeeding protect against antimicrobial resistance?
Maya L. Nadimpalli, Claire D. Bourke, Ruairi C. Robertson, Elisabeth Delarocque-Astagneau, Amee R. Manges, Amy J. Pickering
BMC Medicine.2020;[Epub] CrossRef - The Epidemiological Pattern and Risk Factor of ESBL (Extended Spectrum Β-Lactamase) Producing Enterobacteriaceae in Gut Bacterial Flora of Dairy Cows and People Surrounding in Rural Area, Indonesia
Agusta Reny Soekoyo, Sulistiawati Sulistiawati, Wahyu Setyorini, Kuntaman Kuntaman
Indonesian Journal of Tropical and Infectious Dise.2020; 8(3): 144. CrossRef


 PubReader
PubReader Cite
Cite


