Articles
- Page Path
- HOME > Osong Public Health Res Perspect > Volume 15(1); 2024 > Article
-
Original Article
Prevalence, multidrug resistance, and biofilm formation of Vibrio parahaemolyticus isolated from fish mariculture environments in Cat Ba Island, Vietnam -
Kim Cuc Thi Nguyen, 1
 , Phuc Hung Truong, 2
, Phuc Hung Truong, 2 , Hoa Truong Thi, 3
, Hoa Truong Thi, 3 , Xuan Tuy Ho, 1
, Xuan Tuy Ho, 1 , Phu Van Nguyen, 1
, Phu Van Nguyen, 1
-
Osong Public Health and Research Perspectives 2024;15(1):56-67.
DOI: https://doi.org/10.24171/j.phrp.2023.0181
Published online: February 19, 2024
1Institute of Biotechnology, Hue University, Hue, Vietnam
2Faculty of Biotechnology, TNU-University of Sciences, Tan Thinh Ward, Thai Nguyen, Vietnam
3Department of Fisheries, University of Agriculture and Forestry, Hue University, Hue, Vietnam
- Corresponding author: Phu Van Nguyen Institute of Biotechnology, Hue University, Nguyen Dinh Tu Street, Phu Thuong, Hue 530000, Vietnam E-mail: nguyenvanphu@hueuni.edu.vn
© 2024 Korea Disease Control and Prevention Agency.
This is an open access article under the CC BY-NC-ND license (http://creativecommons.org/licenses/by-nc-nd/4.0/).
- 811 Views
- 49 Download
Abstract
-
Objectives
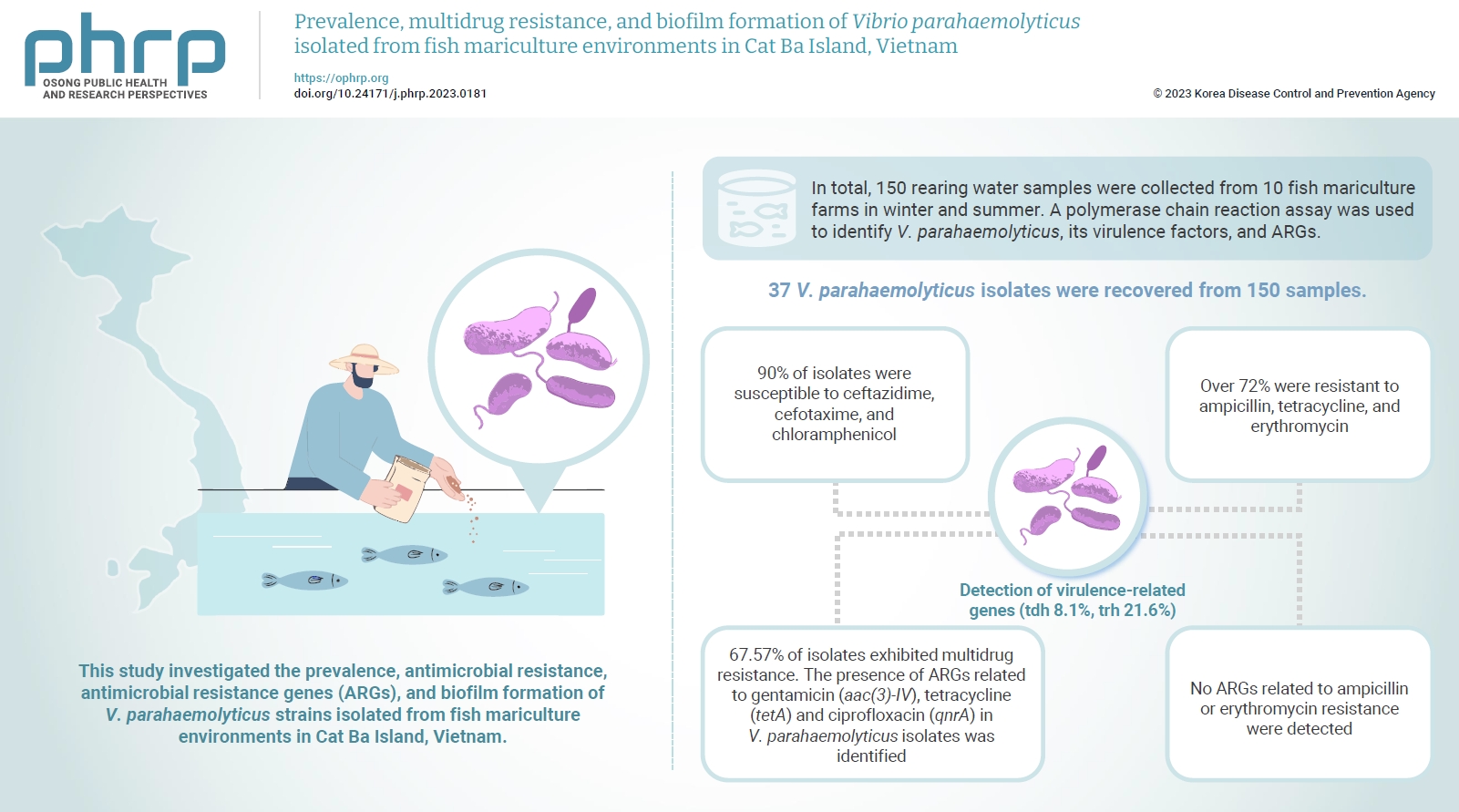
- Vibrio parahaemolyticus is a major foodborne pathogen in aquatic animals and a threat to human health worldwide. This study investigated the prevalence, antimicrobial resistance, antimicrobial resistance genes (ARGs), and biofilm formation of V. parahaemolyticus strains isolated from fish mariculture environments in Cat Ba Island, Vietnam.
-
Methods
- In total, 150 rearing water samples were collected from 10 fish mariculture farms in winter and summer. A polymerase chain reaction assay was used to identify V. parahaemolyticus, its virulence factors, and ARGs. The antimicrobial resistance patterns and biofilm formation ability of V. parahaemolyticus strains were investigated using the disk diffusion test and a microtiter plate-based crystal violet method, respectively.
-
Results
- Thirty-seven V. parahaemolyticus isolates were recovered from 150 samples. The frequencies of the tdh and trh genes among V. parahaemolyticus isolates were 8.1% and 21.6%, respectively. More than 90% of isolates were susceptible to ceftazidime, cefotaxime, and chloramphenicol, but over 72% were resistant to ampicillin, tetracycline, and erythromycin. Furthermore, 67.57% of isolates exhibited multidrug resistance. The presence of ARGs related to gentamicin (aac(3)-IV), tetracycline (tetA) and ciprofloxacin (qnrA) in V. parahaemolyticus isolates was identified. Conversely, no ARGs related to ampicillin or erythromycin resistance were detected. Biofilm formation capacity was detected in significantly more multidrug-resistant isolates (64.9%) than non-multidrug-resistant isolates (18.9%).
-
Conclusion
- Mariculture environments are a potential source of antibiotic-resistant V. parahaemolyticus and a hotspot for virulence genes and ARGs diffusing to aquatic environments. Thus, the prevention of antibiotic-resistant foodborne vibriosis in aquatic animals and humans requires continuous monitoring.
- Aquaculture environments are considered to be reservoirs of various biological pollutants, including infectious agents and antimicrobial resistance genes (ARGs) [1]. ARGs, which are contaminants of emerging concern [2], encode antimicrobial resistance and play an essential role in the process of resistance prevalence and proliferation [3]. In the environment, ARGs can be transferred from one microorganism to another, including from non-pathogenic to pathogenic species, through horizontal gene transfer. Pathogenic microorganisms that carry ARGs within aquaculture settings represent one of the most significant global health hazards, with millions of individuals succumbing to foodborne and waterborne diseases annually [4].
- Vibrio parahaemolyticus is a halophilic marine bacterium that belongs to the family Vibrionaceae and is widely distributed in brackish water and marine environments [5]. This bacterium is a facultative human pathogen and is responsible for approximately 25% of all foodborne diseases closely associated with the consumption of raw seafood [6]. The ingestion of contaminated raw fish and shellfish, or seafood that has not been sufficiently heat-treated, can lead to symptoms such as diarrhea, abdominal pain, vomiting, chills, and low-grade fever in humans [7]. The significance of infections caused by this pathogen is increasing in public health due to a steady rise in its incidence worldwide over recent decades. V. parahaemolyticus is considered an emerging pathogen with a global distribution. Its infectious capability is mediated by various virulence factors, including hemolysin, urease, 2 type III secretion systems, and 2 type VI secretion systems, as well as the formation of persister cells [8]. To survive during infections and in seafood, V. parahaemolyticus cells may also form biofilms, which shield the cells from host defenses and external factors, including antibiotics, thereby contributing to antimicrobial resistance.
- Antibiotics are widely used in aquaculture for both disease prevention and the treatment of infections potentially caused by V. parahaemolyticus [4]. However, the long-term use of antibiotics, particularly at incorrect dosages, can lead to the development of antimicrobial resistance in marine bacteria, including V. parahaemolyticus [4]. Moreover, antibiotics present in aquatic products and seafood can be ingested by animals and humans, potentially leading to the development of resistance in microorganisms that cause diseases in both animals and humans. This resistance can diminish the effectiveness of these antibiotics in treating infections.
- To date, hundreds of V. parahaemolyticus strains have been isolated from seafood and shrimp farming environments in Vietnam [9–11]. However, no previous study has investigated the molecular resistance mechanisms and biofilm formation of V. parahaemolyticus in marine fish environments in Vietnam. Cat Ba Island is a significant marine fish production area and a well-known tourist destination in the northeast of Vietnam. To mitigate outbreaks caused by the consumption of raw or undercooked fish contaminated with V. parahaemolyticus, and to prevent potential risks to human health, it is necessary to monitor the prevalence of V. parahaemolyticus in fish mariculture rearing water. Additionally, identifying antimicrobial resistance and ARGs, virulence factor genes, and biofilm formation is of paramount importance. The aim of this study was to improve our understanding of the current state of antimicrobial resistance in the marine environment and the risks posed by the virulence factor genes and ARGs of V. parahaemolyticus, which can cause foodborne diseases in aquatic animals and humans.
Introduction
- Sampling
- The seawater samples were collected following the method described by Zhao et al. [12], with slight modifications. In brief, water from the fish mariculture environment was collected from a depth of 10 to 20 cm below the seawater surface. A total of 150 samples were collected from 10 grouper mariculture farms in Lan Ha Bay (Cat Ba Island), Hai Phong (Figure S1 and Table S1), during the survey period between February 2021 and August 2021.
- Bacteria Isolation
- V. parahaemolyticus was isolated as described by Mok et al. [13]. Briefly, 5 mL of seawater was added to a flask containing 95 mL of peptone water with 1.5% NaCl and then incubated at 28 °C for 24 hours. Subsequently, 100 μL aliquots were spread on thiosulfate-citrate-bile salts-sucrose (TCBS) agar (Merck) and incubated for 48 hours at 28 °C. Colonies that grew on TCBS were characterized by their color, shape, and size. The blue colonies were subsequently inoculated onto CHROMagar medium (Titan). Purple presumptive colonies were further confirmed through biochemical and polymerase chain reaction (PCR) assays.
- Genomic DNA Extraction and Identification of V. parahaemolyticus
- Colonies of each suspected isolate were cultured in TSB medium (Merck) supplemented with 1.5% NaCl at 28 °C. After 24 hours of incubation, 1.5 mL of the bacterial culture was harvested by centrifugation at 13,000 rpm for 5 minutes. The total genomic DNA from each suspected isolate was then extracted using the Favorgen Tissue Genomic DNA Extraction Mini Kit (Favorgen), following the manufacturer’s instructions. V. parahaemolyticus was identified by a PCR assay using toxR-specific primers [14]. The genomic DNA of V. parahaemolyticus from the collection of the Department of Fisheries, University of Agriculture and Forestry, Hue University was used as a positive control, while nuclease-free water was used as a negative control.
- The presence of virulence genes was identified using the primers listed in Table S2. PCR reactions were performed in PTC200 thermal cyclers (Marshall Scientific). Each 25-µL reaction mixture contained 12.5 µL of MyTaq PCR master mix (Bioline), 1 µL of DNA template (50 ng/µL), 1 µL of each primer (10 pmol), and 9.5 µL of distilled water. The PCR program was as follows: initial denaturation at 95 °C for 5 minutes, followed by 35 cycles of 95 °C for 30 seconds, an annealing temperature of 50 to 60 °C for 30 seconds (varying based on the primer used), and extension at 72 °C for 1 minute, with a final elongation step at 72 °C for 5 minutes. PCR products were visualized by agarose gel electrophoresis using Safe DNA stain (AAT Bioquest).
- Antimicrobial Susceptibility Profiling and PCR Detection of ARGs
- The antimicrobial susceptibility of each V. parahaemolyticus isolate was assessed using the disc diffusion method, following the Clinical and Laboratory Standards Institute (CLSI) M45 guidelines [15]. Initially, colonies from each isolate were suspended in 1% NaCl and adjusted to match the 0.5 McFarland standard. This suspension was then spread onto Mueller-Hinton agar plates (HiMedia). Subsequently, discs impregnated with 9 antimicrobial agents (Mast Diagnostics)—ampicillin (10 μg), ciprofloxacin (5 μg), kanamycin (30 μg), gentamicin (10 μg), tetracycline (30 μg), chloramphenicol (30 μg), erythromycin (15 μg), ceftazidime (30 μg), and ceftiofur (30 μg)—were placed equidistantly on the surface of the agar. The plates were then incubated at 28 °C for 18 to 24 hours. Escherichia coli ATCC 25922 served as the quality control strain. The resistance profile of V. parahaemolyticus was determined by measuring the zones of inhibition, in accordance with the CLSI M45 guidelines for Vibrio spp. and the CLSI M100 guidelines [15,16]. The multiple antibiotic resistance (MAR) index was calculated using the formula a/b, where “a” is the number of antibiotics to which resistance was detected in a single isolate, and “b” represents the total number of antibiotics tested [17].
- Concurrent with antimicrobial resistance testing, genotype screening was performed using PCR assays to detect the presence of ARGs for ampicillin, gentamicin, ciprofloxacin, erythromycin, and tetracycline. The primers for the targeted ARGs and the PCR conditions are detailed in Table S1.
- Biofilm Formation Assay
- The biofilm production capability of V. parahaemolyticus isolates was assessed using a previously described method [18], with some modifications. In brief, overnight cultures of V. parahaemolyticus grown in TSB with 1.5% (w/v) NaCl at 30 °C were diluted to an optical density at 600 nm (OD600) of 0.1. Then, 150 μL of the diluted cell suspension was aliquoted in triplicate into 96-well plates (SPL Life Science). After 24 hours of incubation, non-adherent cells and culture medium were removed, and the attached cells were washed 3 times with a 2 mM CaCl2/MgCl2 buffer. The cells were then stained with 0.01% crystal violet (Merck) for 15 minutes. Subsequently, the plates were washed 3 more times with the 2 mM CaCl2/MgCl2 buffer. To facilitate the measurement of absorbance, 70% ethanol was added to each well before the contents were transferred to a new 96-well plate. Absorbance was measured at a wavelength of 540 nm. The criteria for interpreting the biofilm formation capacity of V. parahaemolyticus were as follows, based on a previous report [19]. Briefly, OD>4×ODc means strong biofilm formation; 2×ODc<OD≤4×Odc, moderate biofilm formation; ODc<OD≤2×Odc, weak biofilm formation; and negative biofilm formation when OD<ODc. ODc refers to the control measurement, which was performed in a microtiter plate without cells.
- Statistical Analysis
- The statistical analysis was carried out using IBM SPSS ver. 20.0 (IBM Corp.). The significance of differences in biofilm formation, the prevalence of V. parahaemolyticus, and the distribution of ARGs was examined using the chi-square test. A p-value ≤0.05 was considered as significant.
Materials and Methods
- Prevalence of V. parahaemolyticus in Fish Farming Water
- The distribution of V. parahaemolyticus in farming water is presented in Table 1. In total, 51 suspected V. parahaemolyticus isolates were obtained from a total of 150 samples, using TCBS agar and CHROMagar. These isolates were further verified by a PCR assay (Figure 1A) with toxR primers, confirming that 37 of them were V. parahaemolyticus. Of these, 11 isolates were collected from 76 samples obtained in the winter, while 26 were identified from 74 samples obtained in the summer. Consequently, the isolation rate of V. parahaemolyticus was significantly higher in the summer (35.1%) than in the winter (14.5%) (p<0.05, chi-square test).
- Detection of Virulence-Related Genes
- The presence of the tdh and trh genes in all 37 V. parahaemolyticus isolates was examined. The results, as shown in Figure 1 and Table 2, indicated that 3 (8.1%) isolates were positive for the tdh gene and 8 (21.6%) isolates were positive for the trh gene. However, none of the isolates carried both the tdh and trh genes. Additionally, biofilm-associated genes were highly prevalent, with 83.8% of isolates harboring the VP0952 gene, and the VP0950 and VP0962 genes were present in 81.08% of isolates.
- Antimicrobial Resistance Pattern of the V. parahaemolyticus Isolates
- The antibiotic resistance profile of 37 isolates against 9 antibiotics is presented in Table 3. None of the 37 isolates exhibited resistance to ceftazidime, cefotaxime, or chloramphenicol. A lower resistance rate was observed for ciprofloxacin (18.9%, 7/37), kanamycin (13.5%, 5/37), and gentamicin (10.8%, 4/37). In contrast, a majority of the isolates were resistant to ampicillin (86.5%, 32/37), erythromycin (75.7%, 28/37), and tetracycline (73.0%, 27/37).
- The multidrug resistance profile of 37 V. parahaemolyticus strains was also examined (Table 2). All 37 V. parahaemolyticus isolates were found to be resistant to at least 1 antibiotic, among which 2 (5.4%), 10 (27.0%), 16 (43.2%), 7 (18.9%), and 2 (5.4%) isolates were resistant to 1, 2, 3, 4, and 5 antibiotics, respectively. Overall, the MAR index ranged from 0.11 to 0.55 (Table 2), with 18 different resistance patterns. The highest frequencies of multidrug-resistant phenotypes were observed for ampicillin, tetracycline, and erythromycin (AMP TCN ERY) with 24.32%. Additionally, 10.81% and 8.1% of isolates exhibited the (AMP TCN) and (AMP GEN TCN ERY) as well as (AMP CIP TCN ERY) phenotypes, respectively.
- ARGs in V. parahaemolyticus Isolates
- The prevalence of 8 ARGs in V. parahaemolyticus isolates was examined. As shown in Table 4, aac(3)-IV, qnrA, and tetA were detected in 16.2%, 27.0%, and 62.2% of isolates, whereas blaTEM, blaOXA, blaSHV, ermA, and ermB were undetected in all isolates tested. Six isolates were aac(3)-IV-positive, 4 were susceptible to gentamicin, and 2 had intermediate resistance to gentamicin. All 10 qnrA-positive isolates were resistant to ciprofloxacin. Of 27 isolates exhibiting resistance to tetracycline, 23 isolates harbored the tetA gene.
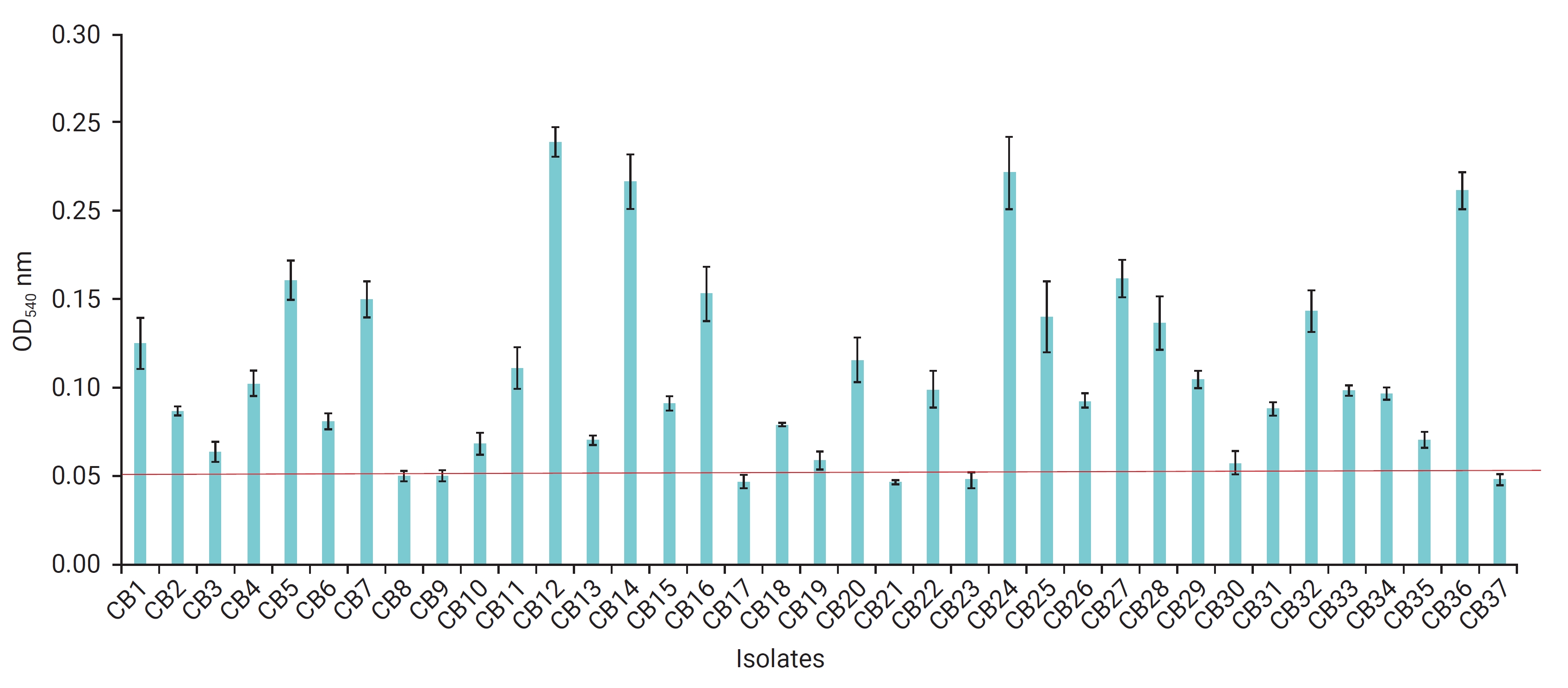
- Biofilm Formation of V. parahaemolyticus Isolates
- The ability of V. parahaemolyticus isolates to form biofilms was investigated, as depicted in Figure 2. Out of the isolates tested, 31 (83.8%) were capable of producing biofilms, whereas 6 (16.2%) could not form biofilms. Within the group of 25 multidrug-resistant isolates, 4 were identified as strong biofilm producers, adhering robustly to polystyrene. Additionally, 11 isolates were classified as moderate biofilm producers, 9 as weak biofilm producers, and only 1 isolate (VA37) was unable to produce biofilm. Conversely, among the 12 non-multidrug-resistant isolates, a single isolate demonstrated moderate biofilm formation, 6 were weak biofilm producers, and 5 were incapable of forming biofilms.
Results
- V. parahaemolyticus is an opportunistic pathogen and a major causative agent of food-borne illnesses worldwide [7]. This bacterium causes gastroenteritis following the consumption of contaminated raw or inadequately cooked seafood. Additionally, exposure to water containing V. parahaemolyticus can lead to wound infections and septicemia, which can be life-threatening for individuals with certain medical conditions, such as diabetes or immune deficiency [7]. The wide distribution of V. parahaemolyticus in tropical seawater and mariculture environments in different countries has been reported [20–23] and its presence depends on the water temperature [13]. In this study, V. parahaemolyticus was present in 24.7% (37/150) of samples, and it was more frequently identified in summer (35.1%) than in winter (14.5%). Previous studies have also noted a higher prevalence of V. parahaemolyticus in summer and autumn than in winter and spring [13,24]. This finding supports the hypothesis that the presence of V. parahaemolyticus is associated with a seasonal pattern, with outbreaks in the warmer season and lower prevalence in the cold season [25]. Since V. parahaemolyticus is cold-susceptible, its growth may be inhibited at lower temperature in winter [7], while higher temperatures in summer promote V. parahaemolyticus growth with a shorter generation time and a faster growth rate, leading to a higher density of V. parahaemolyticus in samples [26].
- It is well known that not all V. parahaemolyticus strains cause diseases in aquatic animals and humans, but strains harboring the tdh and trh genes produce hemolysin factors that induce inflammatory gastroenteritis, and these genes are considered as virulent indicators of pathogenic strains [5]. Therefore, detecting these genes in isolates is crucial for mitigating potential risks to human health. In this study, we identified a significantly higher prevalence of the trh gene (21.6%) than the tdh gene (8.1%) among 37 V. parahaemolyticus isolates. This pattern aligns with previous research, which found the trh and tdh genes in 15.9% and 6.1% of seawater isolates from Korea [25] and in 19.8% and 9.9% of isolates from seafood in China [27], respectively. In contrast, the tdh gene was detected at a higher level (48%) than the trh gene (8.3%) in an estuarine system in South Carolina, the United States in a study by Gutierrez et al. [28] in 2013. Similarly, tdh-positive V. parahaemolyticus was also more prevalent than trh-positive bacteria in oyster environments in Taiwan [29] and in coastal water in Saudi Arabia [30]. The differences in the distribution of tdh+/trh+ V. parahaemolyticus strains are possibly due to the sampling techniques, the geographical origin, seasonal effects and the method of their detection [12]. For example, Parveen et al. [31] reported that real-time PCR could help increase the detection of tdh- and trh-positive V. parahaemolyticus to 13% and 40%, respectively, for water samples compared to the conventional techniques.
- The antimicrobial resistance of Vibrio species has emerged as one of the most significant threats to fish farming, food safety, and public health [32]. Therefore, monitoring the antimicrobial susceptibility of V. parahaemolyticus is very important for evaluating its potential effects on environmental and human health. Our results indicated that more than 72% of V. parahaemolyticus isolates from mariculture were resistant to ampicillin, erythromycin, and tetracycline. These findings broadly support the work of other studies, in which 86% to 100% of V. parahaemolyticus strains isolated from marine environments [33,34] were resistant to ampicillin and tetracycline, and 42% to 48.3% of isolates from shrimp mariculture were resistant to erythromycin [12,35]. This high frequency of resistance confirms that these antimicrobials are widely used and becoming less effective against V. parahaemolyticus, likely due to the extensive use of antibiotics in the areas studied. In Vietnam, 64% of fish farms reported using at least 1 antibiotic for disease treatment and prevention, with 10% to 21% of farms utilizing tetracycline [36]. Additionally, significant levels of antibiotic residues, including ampicillin, erythromycin, and tetracycline, have been detected in aquaculture water in Vietnam, which may contribute to the increasing resistance rates to these antimicrobials [37]. Conversely, other studies have shown that ampicillin, tetracycline, and erythromycin were effective against V. parahaemolyticus strains, with over 90% of isolates being inhibited [10,38–40]. The variability in resistance of Vibrio to antibiotics may be attributed to geographical differences or variations in testing methodologies [5].
- Significantly, all tested V. parahaemolyticus isolates were found to be susceptible to third-generation cephalosporins (ceftazidime and cefotaxime) and chloramphenicol. However, 2 isolates showed intermediate resistance to cefotaxime, and 3 isolates exhibited intermediate resistance to chloramphenicol. These findings are consistent with those of previous studies [12,13,39,41], which reported that over 90% of V. parahaemolyticus strains were susceptible to these antibiotics. Therefore, these drugs are considered the most effective antimicrobials for treating V. parahaemolyticus infections in the areas studied.
- Ciprofloxacin, a fluoroquinolone antibiotic, has been broadly employed as an alternative treatment for tetracycline-resistant bacteria [33], while aminoglycosides (kanamycin and gentamicin) antibiotics are commonly used in aquaculture production and show substantial effectiveness against a broad spectrum of bacteria [42]. However, the use of these antibiotics for the prevention and treatment of V. parahaemolyticus may not be effective, as the rate of antibiotic resistance among 37 V. parahaemolyticus isolates in the current study ranged from 13.5% to 27.0%. Comparable resistance rates have been observed in previous studies, with 9.5% to 39.8% of V. parahaemolyticus strains showing resistance to kanamycin, ciprofloxacin, and gentamicin [24,43]. The increasing resistance of V. parahaemolyticus to fluoroquinolones and aminoglycosides could be attributed to their extensive use in human medicine and aquaculture production, which may lead to significant public health concerns [44].
- MAR indexing has been recognized as an efficient and cost-effective method for tracking the sources of bacterial contamination. Letchumanan et al. [5] suggest that MAR index values greater than 0.2 indicate high-risk sources of antibiotic contamination, posing a potential threat to human health. Regrettably, our study revealed that 67.57% of V. parahaemolyticus isolates had a MAR index exceeding 0.2, indicating a multidrug resistance phenotype. Our findings suggest that these V. parahaemolyticus strains were recovered from sources where antibiotics are frequently used, which could be hazardous. This is in contrast to the findings of Mok et al. [13], who reported that only 1.9% of V. parahaemolyticus strains isolated from the Korean coast were resistant to 3 antibiotics. Nonetheless, our results are in general agreement with previous observations [6,24,45], which showed that more than half of the environmental V. parahaemolyticus isolates exhibited multidrug resistance. Furthermore, the MAR index values in this study ranged from 0.11 to 0.55, which are significantly lower than those reported by Ahmed et al. [46], who found all V. parahaemolyticus isolates were resistant to at least 7 antimicrobial agents. The variation in the MAR index values may be attributed to differences in sample sources, geographic distribution, the number and types of antibiotics tested, and the methodologies employed [47].
- The presence of ARGs is the basis for bacterial resistance, and a high detection rate of ARGs indicates an elevated risk of ARG transmission and a significant potential for bacteria to develop resistance [48]. In the present study, the qnrA gene was detected in all 10 ciprofloxacin-resistant isolates, demonstrating a strong correlation between phenotypic resistance and genotype (the presence of qnrA). This result aligns with previous research [44,45], in which all fluoroquinolone-resistant isolates carried the qnrA and qnrS genes. Conversely, Jeamsripong et al. [49] showed that 77.8% of 594 V. parahaemolyticus isolates harbored the qnr gene, but all isolates were susceptible to ciprofloxacin. This difference is possibly due to the evolution, mutation, and silencing of resistance genes [50] or unknown mechanisms that remain to be elucidated.
- Similarly, a strong relationship between the tetracycline resistance phenotypes of V. parahaemolyticus isolates and the presence of ARG was also determined, as tetA was detected in 23 of 27 isolates that exhibited resistance to tetracycline. The overall level was found to be much higher than reported in other studies [45,51] in which the detection rate of tetA was 28% to 30% of tetracycline-resistant isolates. Meanwhile, tetA was undetected in all V. parahaemolyticus strains that exhibited resistance to tetracycline [5,52]. A possible explanation for this difference might be that V. parahaemolyticus strains used other ARGs (tetM and tetS) against tetracycline [53].
- Interestingly, a high percentage of isolates showed resistance to ampicillin and/or erythromycin; however, none of the 32 or 28 isolates resistant to ampicillin/erythromycin carried the genes for macrolide or β-lactam antibiotic resistance. This discrepancy between the resistance phenotypes and genotypes for ampicillin and/or erythromycin has been noted in previous studies [5,12,45]. This may occur because resistance phenotypes can be governed by various ARGs and mechanisms [12]. Another potential explanation is that the resistance phenotypes are regulated by the efflux systems of the bacterial cells [54].
- The aac(3)-IV gene was detected in susceptible and intermediate-resistance isolates, but not in gentamicin-resistant strains. Similarly, Beshiru and Igbinosa [45] only found the aac(3)-IV gene in 2 intermediate gentamicin-resistant isolates. However, the findings of the current study do not support the previous research [12], in which positive results were found for the aac(3)-IV gene in 12 of 61 isolates that exhibited resistance to gentamicin. Nevertheless, susceptible isolates harbored the aac(3)-IV gene, posing a potential risk for the preservation and transmission of ARGs [12].
- Biofilm formation is a significant virulence factor for pathogenic bacteria during infection. Several Vibrio species, including V. parahaemolyticus, can produce biofilms, enabling the bacteria to establish infections and enhance their resistance to hostile environments, such as antibiotics and the host immune response [55]. This study found that more than 83% of V. parahaemolyticus isolates were biofilm producers (Figure 2). The rate observed in this investigation was 16% lower than that of previous studies, in which 100% of V. parahaemolyticus strains isolated from aquatic animals and marine environments [47,56,57] were able to produce biofilms. The difference in the capacity to form biofilm may be due to the physical conditions (temperature, pH, etc.), and the surfaces where cells attach [58]. Another important finding is that isolates with multidrug resistance (64.9%, 24/37) were more likely to produce biofilms than non-multidrug-resistant isolates (18.9%, 7/37). This supports the possibility that biofilm formation is associated with increased antimicrobial resistance, as suggested in the literature [59–61]. Moreover, the trh gene was detected in 6 biofilm-producing strains that were resistant to 4 or 5 antibiotics. This outcome is consistent with Letchumanan’s findings [62], which indicated that 9 out of 13 trh-positive strains exhibited resistance to 4 or more antimicrobials. Interestingly, a correlation between biofilm phenotype and associated genes was also identified in this study, as 3 genes (VP0950, VP0952, and VP0962) that mediate biofilm formation were present in V. parahaemolyticus isolates that were identified as biofilm producers. Similar results were also reported in other studies [47,56].
- The paper presents intriguing findings on the prevalence of ARGs and the virulence genes of V. parahaemolyticus strains in aquaculture environments and their implications for public health. However, this study did not assess the prevalence of V. parahaemolyticus in fish, which could pose a direct threat to human health. Nevertheless, it is widely recognized that V. parahaemolyticus is an adept swimmer capable of attaching to aquatic animals [63]. Thus, a contaminated water body may potentially infect all its fish inhabitants [64]. Alarmingly, some V. parahaemolyticus isolates identified in the current study carry ARGs and virulence genes, which could pose a significant risk to human health. Aquatic ecosystems, particularly, are acknowledged as hotspots for the environmental spread of pathogenic microorganisms, ARGs, and antimicrobials [1]. Furthermore, the intricate environmental conditions of tropical aquatic habitats may prompt genetic differentiation and adaptive variation in organisms. These changes can enhance their pathogenicity or lead to the emergence of new strains with distinct virulence factors in aquatic hosts [32]. Therefore, to better understand the genotypic variation of V. parahaemolyticus isolates, to determine the relatedness between clinical and environmental isolates, and to clarify the relationships of V. parahaemolyticus virulence genes across different marine environmental sources, further research is required.
Discussion
- This study provides the first insights into the prevalence, antimicrobial resistance profile, distribution of resistance genes, and virulence genes of V. parahaemolyticus strains isolated from fish mariculture environments in Vietnam. The findings suggest that rearing water is a potential source of antimicrobial-resistant V. parahaemolyticus strains, posing a risk to both aquatic animal and human health. The high frequency of resistance to several antibiotics, including ampicillin, erythromycin, and tetracycline, as well as the presence of multidrug-resistant isolates, points to the extensive use of these antibiotics in the region, which should be strictly controlled to prevent their potential spread to humans. Notably, the capacity of several multidrug-resistant isolates to produce strong biofilms indicates the persistence of V. parahaemolyticus strains in marine environments. Therefore, continuous monitoring of V. parahaemolyticus and ARGs in seafood and the mariculture environment is crucial to mitigate potential risks to human health.
Conclusion
- • This is the first study on Vibrio parahaemolyticus isolated from fish mariculture in Vietnam.
- • 8.1% and 21.6% of isolates harbored the tdh and trh genes.
- • A high percentage of isolates exhibited resistance to ampicillin (86.5%), erythromycin (75.7%), and tetracycline (73.0%), and more than 67% of isolates were multidrug-resistant.
- • A close correlation between the resistance phenotypes of V. parahaemolyticus and genotype was identified for ciprofloxacin and tetracycline.
- • Biofilm formation ability was detected in more multidrug-resistant isolates (64.9%) than non-multidrug-resistant isolates (18.9%).
HIGHLIGHTS
Supplementary Material
Figure S1
Table S2.
-
Ethics Approval
Not applicable.
-
Conflicts of Interest
The authors have no conflicts of interest to declare.
-
Funding
This study was supported by the Ministry of Science and Technology of Vietnam with grant number ĐTĐLCN.95/21 and by Hue University under the Core Research Program, Grant No. NCM.DHH2020.13.
-
Availability of Data
All data generated or analyzed during this study are included in this published article (and its Supplementary Information files). For other data, these may be requested through the corresponding author.
-
Authors’ Contributions
Conceptualization: PVN, KCTN; Data curation: PHT, HTT, XTH; Formal analysis: KCTN; Funding acquisition: KCTN; Investigation: all authors; Writing–original draft: PVN, KCTN; Writing–review & editing: all authors. All authors read and approved the final manuscript.
-
Acknowledgements
The authors thank Dr. Derek Wilkinson for proofreading the manuscript.
Article information

| Type of samples | No. of samples | No. of isolates (%) | X2/p* |
|---|---|---|---|
| Sampled in winter | 76 | 11 (14.5) | 8.613/0.003 |
| Sampled in summer | 74 | 26 (35.1) | |
| Total samples | 150 | 37 (24.7) |
- 1. Amarasiri M, Sano D, Suzuki S. Understanding human health risks caused by antibiotic resistant bacteria (ARB) and antibiotic resistance genes (ARG) in water environments: current knowledge and questions to be answered. Crit Rev Environ Sci Technol 2020;50:2016−59.Article
- 2. Pruden A, Pei R, Storteboom H, et al. Antibiotic resistance genes as emerging contaminants: studies in northern Colorado. Environ Sci Technol 2006;40:7445−50.ArticlePubMed
- 3. Dong P, Cui Q, Fang T, et al. Occurrence of antibiotic resistance genes and bacterial pathogens in water and sediment in urban recreational water. J Environ Sci (China) 2019;77:65−74.ArticlePubMed
- 4. Elmahdi S, DaSilva LV, Parveen S. Antibiotic resistance of Vibrio parahaemolyticus and Vibrio vulnificus in various countries: a review. Food Microbiol 2016;57:128−34.ArticlePubMed
- 5. Letchumanan V, Yin WF, Lee LH, et al. Prevalence and antimicrobial susceptibility of Vibrio parahaemolyticus isolated from retail shrimps in Malaysia. Front Microbiol 2015;6:33. ArticlePubMedPMC
- 6. Mok JS, Ryu A, Kwon JY, et al. Distribution of Vibrio species isolated from bivalves and bivalve culture environments along the Gyeongnam coast in Korea: virulence and antimicrobial resistance of Vibrio parahaemolyticus isolates. Food Control 2019;106:106697. Article
- 7. Su YC, Liu C. Vibrio parahaemolyticus: a concern of seafood safety. Food Microbiol 2007;24:549−58.ArticlePubMed
- 8. Guo D, Yang Z, Zheng X, et al. Thymoquinone inhibits biofilm formation and attachment-invasion in host cells of Vibrio parahaemolyticus. Foodborne Pathog Dis 2019;16:671−8.ArticlePubMed
- 9. Ha PT, Thi QV, Thuy NP, et al. Multi‐antibiotics resistance phenotype of pathogenic Vibrio parahaemolyticus isolated from acute hepatopancreatic necrosis disease in Litopenaeus vannamei farmed in the Mekong Delta. J World Aquac Soc 2023;54:1071−87.ArticlePDF
- 10. Vu TT, Hoang TT, Fleischmann S, et al. Quantification and antimicrobial resistance of Vibrio parahaemolyticus in retail seafood in Hanoi, Vietnam. J Food Prot 2022;85:786−91.ArticlePubMedPDF
- 11. Tran THT, Yanagawa H, Nguyen KT, et al. Prevalence of Vibrio parahaemolyticus in seafood and water environment in the Mekong Delta, Vietnam. J Vet Med Sci 2018;80:1737−42.PubMedPMC
- 12. Zhao S, Ma L, Wang Y, et al. Antimicrobial resistance and pulsed-field gel electrophoresis typing of Vibrio parahaemolyticus isolated from shrimp mariculture environment along the east coast of China. Mar Pollut Bull 2018;136:164−70.ArticlePubMed
- 13. Mok JS, Cho SR, Park YJ, et al. Distribution and antimicrobial resistance of Vibrio parahaemolyticus isolated from fish and shrimp aquaculture farms along the Korean coast. Mar Pollut Bull 2021;171:112785. ArticlePubMed
- 14. Kim YB, Okuda J, Matsumoto C, et al. Identification of Vibrio parahaemolyticus strains at the species level by PCR targeted to the toxR gene. J Clin Microbiol 1999;37:1173−7.ArticlePubMedPMCPDF
- 15. Jorgensen JH. Methods for Antimicrobial Dilution and Disk Susceptibility Testing of Infrequently Isolated or Fastidious Bacteria. 2nd ed. CLSI guideline M45. Clinical and Laboratory Standards Institute; 2010.
- 16. Humphries R, Bobenchik AM, Hindler JA, et al. Overview of Changes to the Clinical and Laboratory Standards Institute performance standards for antimicrobial susceptibility testing, M100, 31st edition. J Clin Microbiol 2021;59:e0021321.ArticlePubMedPMCPDF
- 17. Krumperman PH. Multiple antibiotic resistance indexing of Escherichia coli to identify high-risk sources of fecal contamination of foods. Appl Environ Microbiol 1983;46:165−70.ArticlePubMedPMCPDF
- 18. Hammer BK, Bassler BL. Quorum sensing controls biofilm formation in Vibrio cholerae. Mol Microbiol 2003;50:101−4.ArticlePubMed
- 19. Stepanovic S, Vukovic D, Hola V, et al. Quantification of biofilm in microtiter plates: overview of testing conditions and practical recommendations for assessment of biofilm production by staphylococci. APMIS 2007;115:891−9.ArticlePubMed
- 20. Bacian C, Verdugo C, Garcia K, et al. Longitudinal study of total and pathogenic Vibrio parahaemolyticus (tdh+ and/or trh+) in two natural extraction areas of Mytilus chilensis in Southern Chile. Front Microbiol 2021;12:621737. ArticlePubMedPMC
- 21. Jiang H, Yu T, Yang Y, et al. Co-occurrence of antibiotic and heavy metal resistance and sequence type diversity of Vibrio parahaemolyticus isolated from Penaeus vannamei at freshwater farms, seawater farms, and markets in Zhejiang province, China. Front Microbiol 2020;11:1294. ArticlePubMedPMC
- 22. Narayanan SV, Joseph TC, Peeralil S, et al. Tropical shrimp aquaculture farms harbour pathogenic Vibrio parahaemolyticus with high genetic diversity and Carbapenam resistance. Mar Pollut Bull 2020;160:111551. ArticlePubMed
- 23. Tey YH, Jong KJ, Fen SY, et al. Occurrence of Vibrio parahaemolyticus, Vibrio cholerae, and Vibrio vulnificus in the Aquacultural Environments of Taiwan. J Food Prot 2015;78:969−76.ArticlePubMedPDF
- 24. Yang JH, Mok JS, Jung YJ, et al. Distribution and antimicrobial susceptibility of Vibrio species associated with zooplankton in coastal area of Korea. Mar Pollut Bull 2017;125:39−44.ArticlePubMed
- 25. Di DY, Lee A, Jang J, et al. Season-specific occurrence of potentially pathogenic Vibrio spp. on the southern coast of South Korea. Appl Environ Microbiol 2017;83:e02680. −16.ArticlePubMedPMCPDF
- 26. Kim YW, Lee SH, Hwang IG, et al. Effect of temperature on growth of Vibrio parahaemolyticus [corrected] and Vibrio vulnificus in flounder, salmon sashimi and oyster meat. Int J Environ Res Public Health 2012;9:4662−75.PubMedPMC
- 27. Li Y, Xie T, Pang R, et al. Food-Borne Vibrio parahaemolyticus in China: prevalence, antibiotic susceptibility, and genetic characterization. Front Microbiol 2020;11:1670. ArticlePubMedPMC
- 28. Gutierrez West CK, Klein SL, Lovell CR. High frequency of virulence factor genes tdh, trh, and tlh in Vibrio parahaemolyticus strains isolated from a pristine estuary. Appl Environ Microbiol 2013;79:2247−52.ArticlePubMedPMCPDF
- 29. Chang HC, Chen ML, Su YC, et al. Molecular characterizations of pathogenic Vibrio parahaemolyticus isolated from Southern Taiwan oyster-growing environment. Food Control 2011;22:245−51.Article
- 30. Almejhim M, Aljeldah M, Elhadi N. Improved isolation and detection of toxigenic Vibrio parahaemolyticus from coastal water in Saudi Arabia using immunomagnetic enrichment. PeerJ 2021;9:e12402.ArticlePubMedPMCPDF
- 31. Parveen S, Hettiarachchi KA, Bowers JC, et al. Seasonal distribution of total and pathogenic Vibrio parahaemolyticus in Chesapeake Bay oysters and waters. Int J Food Microbiol 2008;128:354−61.ArticlePubMed
- 32. Yu Y, Li H, Wang Y, et al. Antibiotic resistance, virulence and genetic characteristics of Vibrio alginolyticus isolates from aquatic environment in costal mariculture areas in China. Mar Pollut Bull 2022;185(Pt A):114219. Article
- 33. Kang CH, Shin Y, Jang S, et al. Characterization of Vibrio parahaemolyticus isolated from oysters in Korea: resistance to various antibiotics and prevalence of virulence genes. Mar Pollut Bull 2017;118:261−6.ArticlePubMed
- 34. Siddique AB, Moniruzzaman M, Ali S, et al. Characterization of pathogenic Vibrio parahaemolyticus isolated from fish aquaculture of the southwest coastal area of Bangladesh. Front Microbiol 2021;12:635539. ArticlePubMedPMC
- 35. Kitiyodom S, Khemtong S, Wongtavatchai J, et al. Characterization of antibiotic resistance in Vibrio spp. isolated from farmed marine shrimps (Penaeus monodon). FEMS Microbiol Ecol 2010;72:219−27.ArticlePubMed
- 36. Luu QH, Nguyen TB, Nguyen TL, et al. Antibiotics use in fish and shrimp farms in Vietnam. Aquac Rep 2021;20:100711. Article
- 37. Binh VN, Dang N, Anh NT, et al. Antibiotics in the aquatic environment of Vietnam: sources, concentrations, risk and control strategy. Chemosphere 2018;197:438−50.ArticlePubMed
- 38. Xie T, Yu Q, Tang X, et al. Prevalence, antibiotic susceptibility and characterization of Vibrio parahaemolyticus isolates in China. FEMS Microbiol Lett 2020;367:fnaa136. ArticlePubMedPDF
- 39. Tan CW, Rukayadi Y, Hasan H, et al. Prevalence and antibiotic resistance patterns of Vibrio parahaemolyticus isolated from different types of seafood in Selangor, Malaysia. Saudi J Biol Sci 2020;27:1602−8.ArticlePubMedPMC
- 40. Park K, Mok JS, Ryu AR, et al. Occurrence and virulence of Vibrio parahaemolyticus isolated from seawater and bivalve shellfish of the Gyeongnam coast, Korea, in 2004-2016. Mar Pollut Bull 2018;137:382−7.ArticlePubMed
- 41. Stratev D, Fasulkova R, Krumova-Valcheva G. Incidence, virulence genes and antimicrobial resistance of Vibrio parahaemolyticus isolated from seafood. Microb Pathog 2023;177:106050. ArticlePubMed
- 42. Pepi M, Focardi S. Antibiotic-resistant bacteria in aquaculture and climate change: a challenge for health in the Mediterranean area. Int J Environ Res Public Health 2021;18:5723. ArticlePubMedPMC
- 43. Igbinosa EO, Beshiru A, Igbinosa IH, et al. Prevalence and characterization of food-borne Vibrio parahaemolyticus from African salad in Southern Nigeria. Front Microbiol 2021;12:632266. ArticlePubMedPMC
- 44. Lei T, Jiang F, He M, et al. Prevalence, virulence, antimicrobial resistance, and molecular characterization of fluoroquinolone resistance of Vibrio parahaemolyticus from different types of food samples in China. Int J Food Microbiol 2020;317:108461. ArticlePubMed
- 45. Beshiru A, Igbinosa EO. Surveillance of Vibrio parahaemolyticus pathogens recovered from ready-to-eat foods. Sci Rep 2023;13:4186. ArticlePubMedPMCPDF
- 46. Ahmed HA, El Bayomi RM, Hussein MA, et al. Molecular characterization, antibiotic resistance pattern and biofilm formation of Vibrio parahaemolyticus and V. cholerae isolated from crustaceans and humans. Int J Food Microbiol 2018;274:31−7.ArticlePubMed
- 47. Ashrafudoulla M, Mizan MF, Park H, et al. Genetic relationship, virulence factors, drug resistance profile and biofilm formation ability of Vibrio parahaemolyticus isolated from mussel. Front Microbiol 2019;10:513. ArticlePubMedPMC
- 48. Dutta D, Kaushik A, Kumar D, et al. Foodborne pathogenic Vibrios: antimicrobial resistance. Front Microbiol 2021;12:638331. ArticlePubMedPMC
- 49. Jeamsripong S, Khant W, Chuanchuen R. Distribution of phenotypic and genotypic antimicrobial resistance and virulence genes in Vibrio parahaemolyticus isolated from cultivated oysters and estuarine water. FEMS Microbiol Ecol 2020;96:fiaa081. ArticlePubMedPMCPDF
- 50. Liu M, Wong MH, Chen S. Mechanisms of fluoroquinolone resistance in Vibrio parahaemolyticus. Int J Antimicrob Agents 2013;42:187−8.ArticlePubMed
- 51. Adesiyan IM, Bisi-Johnson MA, Okoh AI. Incidence of antibiotic resistance genotypes of Vibrio species recovered from selected freshwaters in Southwest Nigeria. Sci Rep 2022;12:18912. ArticlePubMedPMCPDF
- 52. Jiang Y, Yao L, Li F, et al. Characterization of antimicrobial resistance of Vibrio parahaemolyticus from cultured sea cucumbers (Apostichopus japonicas). Lett Appl Microbiol 2014;59:147−54.ArticlePubMedPDF
- 53. Kim SR, Nonaka L, Suzuki S. Occurrence of tetracycline resistance genes tet(M) and tet(S) in bacteria from marine aquaculture sites. FEMS Microbiol Lett 2004;237:147−56.ArticlePubMed
- 54. Pazhani GP, Bhowmik SK, Ghosh S, et al. Trends in the epidemiology of pandemic and non-pandemic strains of Vibrio parahaemolyticus isolated from diarrheal patients in Kolkata, India. PLoS Negl Trop Dis 2014;8:e2815.ArticlePubMedPMC
- 55. De Silva LA, Heo GJ. Biofilm formation of pathogenic bacteria isolated from aquatic animals. Arch Microbiol 2022;205:36. PubMed
- 56. Mizan MF, Bang HJ, Sadekuzzaman M, et al. Molecular characteristics, biofilm-forming abilities, and quorum sensing molecules in Vibrio parahaemolyticus strains isolated from marine and clinical environments in Korea. Biofouling 2017;33:369−78.ArticlePubMed
- 57. Elexson N, Yaya R, Nor AM, et al. Biofilm assessment of Vibrio parahaemolyticus from seafood using Random Amplified Polymorphism DNA-PCR. Int Food Res J 2014;21:59.
- 58. Han N, Mizan MF, Jahid IK, et al. Biofilm formation by Vibrio parahaemolyticus on food and food contact surfaces increases with rise in temperature. Food Control 2016;70:161−6.Article
- 59. Nirwati H, Sinanjung K, Fahrunissa F, et al. Biofilm formation and antibiotic resistance of Klebsiella pneumoniae isolated from clinical samples in a tertiary care hospital, Klaten, Indonesia. BMC Proc 2019;13(Suppl 11):20. ArticlePubMedPMCPDF
- 60. Santajit S, Kong-Ngoen T, Tunyong W, et al. Occurrence, antimicrobial resistance, virulence, and biofilm formation capacity of Vibrio spp. and Aeromonas spp. isolated from raw seafood marketed in Bangkok, Thailand. Vet World 2022;15:1887−95.ArticlePubMedPMCPDF
- 61. Tian C, Yuan M, Tao Q, et al. Discovery of novel resistance mechanisms of Vibrio parahaemolyticus biofilm against aminoglycoside antibiotics. Antibiotics (Basel) 2023;12:638. ArticlePubMedPMC
- 62. Letchumanan V, Chan KG, Lee LH. Vibrio parahaemolyticus: a review on the pathogenesis, prevalence, and advance molecular identification techniques. Front Microbiol 2014;5:705. ArticlePubMedPMC
- 63. McCarter L. The multiple identities of Vibrio parahaemolyticus. J Mol Microbiol Biotechnol 1999;1:51−7.PubMed
- 64. Kim JY, Lee JL. Correlation of total bacterial and Vibrio spp. populations between fish and water in the aquaculture system. Front Mar Sci 2017;4:147. Article
References
Figure & Data
References
Citations



 Cite
Cite