Search
- Page Path
- HOME > Search
Original Article
- Prevalence, multidrug resistance, and biofilm formation of Vibrio parahaemolyticus isolated from fish mariculture environments in Cat Ba Island, Vietnam
- Kim Cuc Thi Nguyen, Phuc Hung Truong, Hoa Truong Thi, Xuan Tuy Ho, Phu Van Nguyen
- Osong Public Health Res Perspect. 2024;15(1):56-67. Published online February 19, 2024
- DOI: https://doi.org/10.24171/j.phrp.2023.0181
- 917 View
- 50 Download
-
 Graphical Abstract
Graphical Abstract
 Abstract
Abstract
 PDF
PDF 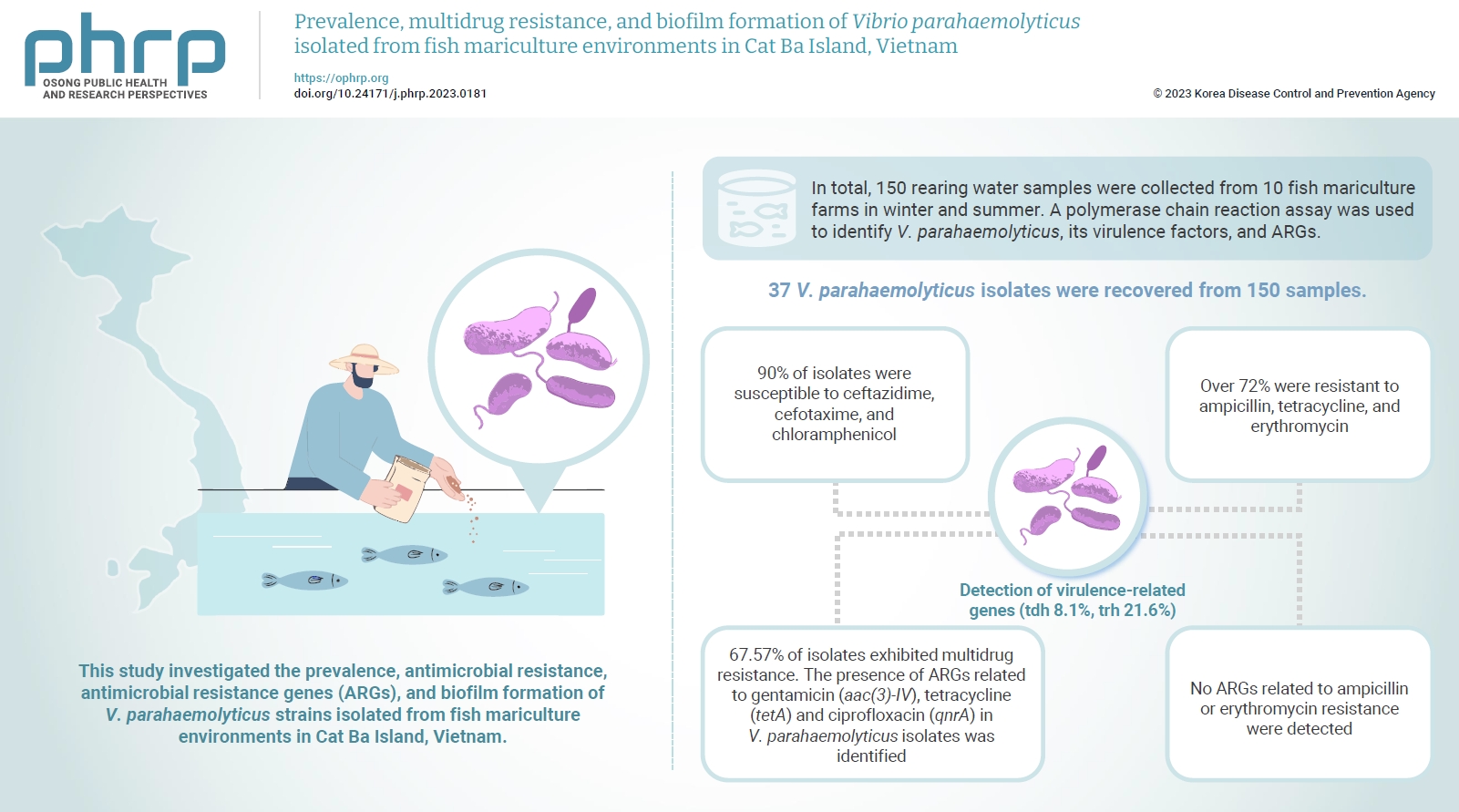
- Objectives
Vibrio parahaemolyticus is a major foodborne pathogen in aquatic animals and a threat to human health worldwide. This study investigated the prevalence, antimicrobial resistance, antimicrobial resistance genes (ARGs), and biofilm formation of V. parahaemolyticus strains isolated from fish mariculture environments in Cat Ba Island, Vietnam. Methods: In total, 150 rearing water samples were collected from 10 fish mariculture farms in winter and summer. A polymerase chain reaction assay was used to identify V. parahaemolyticus, its virulence factors, and ARGs. The antimicrobial resistance patterns and biofilm formation ability of V. parahaemolyticus strains were investigated using the disk diffusion test and a microtiter plate-based crystal violet method, respectively. Results: Thirty-seven V. parahaemolyticus isolates were recovered from 150 samples. The frequencies of the tdh and trh genes among V. parahaemolyticus isolates were 8.1% and 21.6%, respectively. More than 90% of isolates were susceptible to ceftazidime, cefotaxime, and chloramphenicol, but over 72% were resistant to ampicillin, tetracycline, and erythromycin. Furthermore, 67.57% of isolates exhibited multidrug resistance. The presence of ARGs related to gentamicin (aac(3)-IV), tetracycline (tetA) and ciprofloxacin (qnrA) in V. parahaemolyticus isolates was identified. Conversely, no ARGs related to ampicillin or erythromycin resistance were detected. Biofilm formation capacity was detected in significantly more multidrug-resistant isolates (64.9%) than non-multidrug-resistant isolates (18.9%). Conclusion: Mariculture environments are a potential source of antibiotic-resistant V. parahaemolyticus and a hotspot for virulence genes and ARGs diffusing to aquatic environments. Thus, the prevention of antibiotic-resistant foodborne vibriosis in aquatic animals and humans requires continuous monitoring.
Review Article
- Strategies to combat Gram-negative bacterial resistance to conventional antibacterial drugs: a review
- Priyanka Bhowmik, Barkha Modi, Parijat Roy, Antarika Chowdhury
- Osong Public Health Res Perspect. 2023;14(5):333-346. Published online October 18, 2023
- DOI: https://doi.org/10.24171/j.phrp.2022.0323
- 2,269 View
- 177 Download
- 1 Web of Science
- 2 Crossref
-
 Graphical Abstract
Graphical Abstract
 Abstract
Abstract
 PDF
PDF 
- The emergence of antimicrobial resistance raises the fear of untreatable diseases. Antimicrobial resistance is a multifaceted and dynamic phenomenon that is the cumulative result of different factors. While Gram-positive pathogens, such as methicillin-resistant Staphylococcus aureus and Clostridium difficile, were previously the most concerning issues in the field of public health, Gram-negative pathogens are now of prime importance. The World Health Organization’s priority list of pathogens mostly includes multidrug-resistant Gram-negative organisms particularly carbapenem-resistant Enterobacterales, carbapenem-resistant Pseudomonas aeruginosa, and extensively drug-resistant Acinetobacter baumannii. The spread of Gram-negative bacterial resistance is a global issue, involving a variety of mechanisms. Several strategies have been proposed to control resistant Gram-negative bacteria, such as the development of antimicrobial auxiliary agents and research into chemical compounds with new modes of action. Another emerging trend is the development of naturally derived antibacterial compounds that aim for targets novel areas, including engineered bacteriophages, probiotics, metal-based antibacterial agents, odilorhabdins, quorum sensing inhibitors, and microbiome-modifying agents. This review focuses on the current status of alternative treatment regimens against multidrug-resistant Gram-negative bacteria, aiming to provide a snapshot of the situation and some information on the broader context.
-
Citations
Citations to this article as recorded by- Efficacy of new generation biosorbents for the sustainable treatment of antibiotic residues and antibiotic resistance genes from polluted waste effluent
Barkha Madhogaria, Sangeeta Banerjee, Atreyee Kundu, Prasanta Dhak
Infectious Medicine.2024; 3(1): 100092. CrossRef - Evaluation of Plant-Based Silver Nanoparticles for Antioxidant Activity and Promising Wound-Healing Applications
Maria Qubtia, Shazia Akram Ghumman, Sobia Noreen, Huma Hameed, Shazia Noureen, Rizwana Kausar, Ali Irfan, Pervaiz Akhtar Shah, Hafsa Afzal, Misbah Hameed, Mohammad Raish, Maria Rana, Ajaz Ahmad, Katarzyna Kotwica-Mojzych, Yousef A. Bin Jardan
ACS Omega.2024; 9(10): 12146. CrossRef
- Efficacy of new generation biosorbents for the sustainable treatment of antibiotic residues and antibiotic resistance genes from polluted waste effluent
Original Article
- Prevalence of Antimicrobial Resistance in
Escherichia coli Strains Isolated from Fishery Workers - Hyun-Ho Shin, Seung-Hak Cho
- Osong Public Health Res Perspect. 2013;4(2):72-75. Published online April 30, 2013
- DOI: https://doi.org/10.1016/j.phrp.2013.03.001
- 2,906 View
- 16 Download
- 6 Crossref
-
 Abstract
Abstract
 PDF
PDF - Objectives
This study aimed to characterize the prevalence of antibiotic resistance in Escherichia coli isolates from the fecal samples of fishery workers who work in fish farms and often use antibiotics for the feeding fishes.
Methods
Seventy-three E. coli strains isolated from the fecal samples of fishery workers and 180 isolates from a control group of restaurant workers were tested for antibiotic resistance by agar disk diffusion with 16 antimicrobial agents.
Results
About 30% of isolates from each group showed antimicrobial resistance to ampicillin, and 60% of isolates from fishery workers and 41% from restaurant workers were resistant to tetracycline. The isolates showed higher resistance to cephalothin and cefoxitin than to other cephem antibiotics and to gentamicin than to other aminogycosides. Our data indicated that fecal E. coli isolates from fishery workers showed higher antibiotic resistance than those of non-fishery workers (restaurant workers), especially to cephalothin, tetracycline, and trimethoprim–sulfamethoxazole (p < 0.05). However, rates of multidrug resistance were similar among the fishery workers and restaurant workers.
Conclusion
Frequent use of antibiotics may cause increased antibiotic resistance in the human microbiome. -
Citations
Citations to this article as recorded by- Enhanced catalytic performance of Cu2ZnSnS4/MoS2 nanocomposites based counter electrode for Pt-free dye-sensitized solar cells
P. Baskaran, K.D. Nisha, S. Harish, H. Ikeda, J. Archana, M. Navaneethan
Journal of Alloys and Compounds.2022; 894: 162166. CrossRef - Influence of Na2S treatment on CZTS/Mo interface in Cu2ZnSnS4 solar cells annealed in sulfur-free atmosphere
Jie Guo, Shuaihui Sun, Bin Liu, Ruiting Hao, Licun Sun
Optik.2021; 242: 166998. CrossRef - Evaluating antimicrobial resistance in the global shrimp industry
Kelly Thornber, David Verner‐Jeffreys, Steve Hinchliffe, Muhammad Meezanur Rahman, David Bass, Charles R. Tyler
Reviews in Aquaculture.2020; 12(2): 966. CrossRef - Antibiotic-resistant pathogens in the occupational environment
Anna Ławniczek-Wałczyk, Rafał L. Górny
Occupational Safety – Science and Practice.2019; 579(12): 9. CrossRef - Human, animal and environmental contributors to antibiotic resistance in low-resource settings: integrating behavioural, epidemiological and One Health approaches
Emily K. Rousham, Leanne Unicomb, Mohammad Aminul Islam
Proceedings of the Royal Society B: Biological Sci.2018; 285(1876): 20180332. CrossRef - Possibility of CTX-M-14 Gene Transfer from Shigella sonnei to a Commensal Escherichia coli Strain of the Gastroenteritis Microbiome
Seung-Hak Cho, Soon Young Han, Yeon-Ho Kang
Osong Public Health and Research Perspectives.2014; 5(3): 156. CrossRef
- Enhanced catalytic performance of Cu2ZnSnS4/MoS2 nanocomposites based counter electrode for Pt-free dye-sensitized solar cells



 First
First Prev
Prev


