Articles
- Page Path
- HOME > Osong Public Health Res Perspect > Volume 13(6); 2022 > Article
-
Review Article
Carbapenem resistance in critically important human pathogens isolated from companion animals: a systematic literature review -
Angie Alexandra Rincón-Real
 , Martha Cecilia Suárez-Alfonso
, Martha Cecilia Suárez-Alfonso
-
Osong Public Health and Research Perspectives 2022;13(6):407-423.
DOI: https://doi.org/10.24171/j.phrp.2022.0033
Published online: December 16, 2022
Molecular Genetics of Pathogens Group, National University of Colombia, Bogotá, Colombia
- Corresponding author: Martha Cecilia Suárez-Alfonso Molecular Genetics of Pathogens Group, Universidad Nacional of Colombia, Carrera 30 # 45-03, Facultad de Medicina Veterinaria y de Zootecnia Edificio 503, Laboratorio de Microbiología Veterinaria, 111321 Bogotá, Colombia E-mail: mcsuarezal@unal.edu.co
© 2022 Korea Disease Control and Prevention Agency.
This is an open access article under the CC BY-NC-ND license (http://creativecommons.org/licenses/by-nc-nd/4.0/).
Abstract
- This study aimed to describe the presence and geographical distribution of Gram-negative bacteria considered critical on the priority list of antibiotic-resistant pathogens published by the World Health Organization, including carbapenem-resistant Enterobacteriaceae, carbapenem-resistant Acinetobacter spp., and carbapenem-resistant Pseudomonas aeruginosa. A systematic review of original studies published in 5 databases between 2010 and 2021 was conducted, including genotypically confirmed carbapenem-resistant isolates obtained from canines, felines, and their settings. Fifty-one articles met the search criteria. Carbapenem-resistant isolates were found in domestic canines and felines, pet food, and on veterinary-medical and household surfaces. The review found that the so-called “big five”—that is, the 5 major carbapenemases identified worldwide in Enterobacterales (New Delhi metallo-β-lactamase, active-on-imipenem, Verona integron-encoded metallo-β-lactamase, Klebsiella pneumoniae carbapenemase, and oxacillin [OXA]-48-like)—and the 3 most important carbapenemases from Acinetobacter spp. (OXA-23-like, OXA-40-like, and OXA-58-like) had been detected in 8 species in the Enterobacteriaceae family and 5 species of glucose non-fermenting bacilli on 5 continents. Two publications used molecular analysis to confirm carbapenem-resistant bacteria transmission between owners and dogs. Isolating critically important human carbapenem-resistant Gram-negative bacteria from domestic canines and felines highlights the importance of including these animal species in surveillance programs and antimicrobial resistance containment plans as part of the One Health approach.
- A systematic analysis of antimicrobial resistance (AMR) in 2019 found that bacteria-related AMR accounted for 4.5 million deaths worldwide. Moreover, the global burden of AMR will be responsible for 10 million deaths worldwide by 2050 [1]. AMR incurs increased healthcare system costs associated with length of hospital stay, additional follow-up visits, and using drugs of last resort (DoLR) [2−4].
- Antimicrobial-resistant infections in clinical and community settings are frequently associated with β-lactam-resistant Gram-negative bacteria [5−7]. Resistance to β-lactams in Gram-negative bacteria occurs due to target site modification, decreased antibiotic concentration resulting from efflux pumps, changes in outer membrane permeability caused by the loss or modifications of porins, and enzymatic inactivation of the drug by β-lactamase production. More than 4.900 β-lactamases have been reported to date [6,8]. β-lactamases can be structurally [9] or functionally classified [10]. Four types are recognized structurally. Types A, C, and D are serine enzymes, and type B includes metallo-β-lactamases [6,8,9].
- Carbapenemases are extremely relevant because they hydrolyze carbapenems; β-lactam DoLRs have broad-spectrum activity and stability [6]. Carbapenem resistance (CR) is considered a marker for extensively drug-resistant (XDR) and pandrug-resistant (PDR) Gram-negative bacteria because it is associated with a wide range of co-resistance to other antimicrobial drugs [11].
- According to epidemiological factors related to the degree of global spread of carbapenem-hydrolyzing enzymes, 2 groups of carbapenemases have been proposed for Enterobacterales. The first group comprises the “big five” that are widespread worldwide, and the second group includes the minor or “rare” carbapenemases that have a limited geographical spread [8]. The “big five” carbapenemases include the class A enzyme Klebsiella pneumoniae carbapenemase (KPC), class B enzymes active-on-imipenem (IMP), Verona integron-encoded metallo-β-lactamase (VIM), New Delhi metallo-β-lactamase (NDM), and class D enzyme active on oxacillin [OXA]-48-like [8]. Three oxacillinases from the genus Acinetobacter (OXA-23-like, OXA-40-like, and OXA-58-like) have been reported as of concern due to their worldwide spread [12,13].
- The World Health Organization (WHO) published a global priority list of antibiotic-resistant bacteria to guide the discovery, research, and development of new antibiotics in 2017. The most important category on the list (i.e., critical-priority microorganisms) includes the carbapenem-resistant Enterobacteriaceae (CRE) family and 2species of carbapeem-resistant, glucose-non-fermenting bacilli (CRGNFB), namely, carbapenem-resistant Acinetobacter baumannii and carbapenem-resistant Pseudomonas aeruginosa, due to their impact on mortality, disease burden, and circulation at the human-animal-environment interface [11].
- AMR involves complex human-animal-environment interface-related microbial interactions. Carbapenem use is not recommended for companion animals [14]; however, CR isolates have been reported in these animals [15]. The interactions between owners and companion animals promote AMR dissemination and maintenance through bacterial bidirectional transmission [16]. Thus, a One Health-oriented approach to analyzing carbapenemase circulation in companion animals is essential—that is, an integrated, multisector approach seeking to balance and optimize the health of humans and animals, as well as environmental sustainability [17,18].
- Accordingly, this study adopted a One Health perspective for describing the presence and geographic distribution of antibiotic-resistant Gram-negative bacteria classified as critical on the WHO priority list isolated from domestic canines and felines and the contexts associated with their presence, including CRE, carbapenem-resistant A. baumannii, and carbapenem-resistant P. aeruginosa.
Introduction
- Search Strategy and Selection of Studies
- A search for original articles that evaluated antimicrobial susceptibility to carbapenems in Enterobacteriaceae and glucose non-fermenting bacilli (GNFB), such as Acinetobacter spp. and P. aeruginosa obtained from domestic canines or felines, or both, and contexts associated with their presence (e.g., veterinary care, pet food, and/or the animal’s home) in which genotypic CR was detected was carried out. The databases consulted were Medline, PubMed, Web of Science, Scopus, Wiley Online Library, and CABI: VetMed Resource.
- Descriptors from the DeCS/MeSH thesauri were used by applying a specific search formula and combining the following terms: (carbapenemase OR CPE OR carbapenem-resistance) AND (Enterobacteriaceae OR Enterobacterales OR Escherichia coli OR Enterobacter cloacae OR Klebsiella pneumoniae OR Pseudomonas OR Pseudomonas aeruginosa OR Acinetobacter OR Acinetobacter baumannii) AND (companion animals OR pets OR dog OR cat) NOT review (Table S1).
- Articles in English, Spanish or Portuguese, published from January 1, 2010 to April 24, 2021 were included. Publications that informed only about isolates from human sources, animals other than canines or felines (origin or unrelated environmental origin), reports of phenotypic resistance without genotypic confirmation, review articles or meta-analyses, editorials, book chapters, proceedings of academic events, evaluation of diagnostic or therapeutic methods of CR bacteria, were excluded.
- Duplicates were removed using the Mendeley bibliographic manager ver. 1.19.8 (Elsevier, Amsterdam, The Netherlands). Two researchers carried out the search and selection independently in 2 phases. The first phase was based on the title and abstract, and the second was involved analyzing the full text. We conducted a manual search of the reference lists of included articles and selected publications according to the previous criteria to ensure better information coverage. Divergences in selection were resolved by consensus.
- Data Extraction and Processing
- The information was compiled in an Excel sheet (Microsoft Corp., Redmond, WA, USA). It included the main author, year, country, place, sample collected, year of isolation, animal, population, health status, history of hospitalization or antimicrobial therapy in animals and/or owners, as well as the microorganism identified, number of isolates, sequence type (ST), epidemiological classification of AMR, susceptibility assessment, and the genetic mechanism of CR.
Materials and Methods
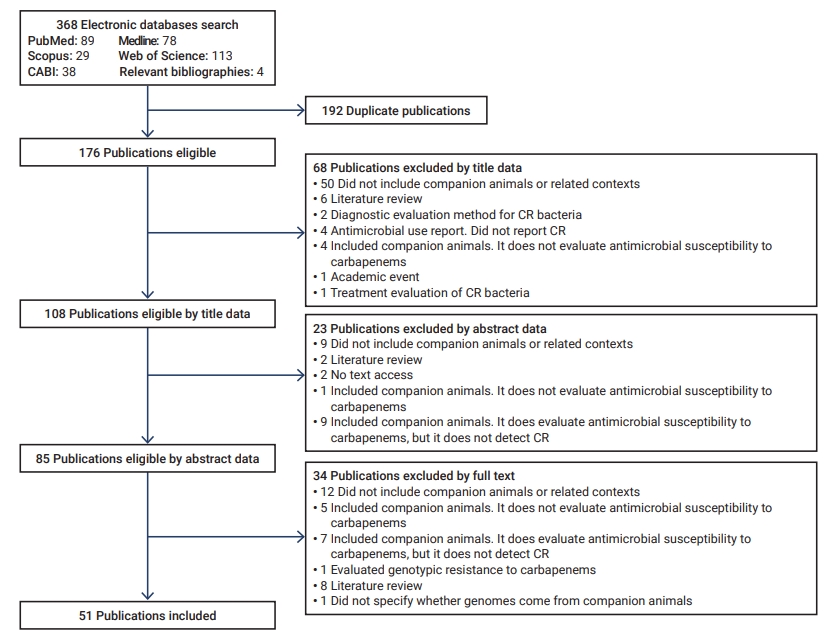
- The initial search yielded 368 articles, 192 of which were duplicates. Of these, 91 were discarded according to their title and abstract, and 34 based on the full-text review (Figure 1). Fifty-one articles were included for analysis [19−69].
- Study Characterization
- Of the 51 articles, 48 reported isolates from domestic canines and/or felines, 3 from veterinary medical care environments, 1 from home environments, and another from commercial food (wet food). Of the 48 publications, 26 included dogs, 4 involved cats, and 18 encompassed both species.
- Furthermore, of the 51 publications, 40 were original studies, and 11 were from epidemiological surveillance/monitoring programs. The publications included articles spanning 11 years (2010−2021); however, some CR isolates reported a longer period of analysis (Table 1) [19−69].
- Animal Clinical History
- Of the 51 articles reporting CRE and CRGNFB, most (n=49) described the health status of animals. Four studies involved healthy animals, 38 included diseased animals, and 5 had animals of both statuses. In the reports of diseased animals, CRE isolates were obtained from different systems, including genitourinary, respiratory, gastrointestinal, cardiovascular, musculoskeletal, ear, skin, and soft tissues, as well as neoplasms and wounds of undescribed origin. The CRGNFB were obtained from respiratory, genitourinary, ocular and ear tissues, soft tissues, and systemic examinations (Table 2; Table S2) [19−69].
- Twenty articles reported antimicrobial therapy close to the sampling date, previously, or both. Three articles reported the administration of carbapenems (meropenem) in South Korea, and 17 described the use of other antimicrobials, including tetracyclines, cephalosporins, and quinolones, β-lactams, β-lactams/β-lactamase inhibitors, aminoglycosides, lincosamides, sulfonamides, nitroimidazoles, and phosphonic acids (Table 2).
- CR Bacteria Isolated from Domestic Canines and Felines
- Twenty publications reported information on the frequency of animals with CR isolates. In dogs and cats, the proportions of CRE isolation ranged from 0.25% to 21.6%. The frequency of CRE was registered on 3 continents. In Europe, the highest frequency of CRE happened in an outbreak of CR E. coli in dogs in a veterinary hospital in Switzerland (21.65%). In Asia, the highest frequency of CRE was in dogs in veterinary hospitals in India (6.75%). In Africa, the highest frequency of CRE occurred in animals sampled at an official veterinary office in Algeria (2.5%) (Table 3) [19,20,24−28,30−33,44,45,48,56,59,61,63,67].
- The frequency of companion animals from CRGNFB isolates ranged from 1.3% to 12.50%. The frequency of CRGNFB was reported on 2 continents. In Asia, the highest frequency of CRGNFB was registered in dogs in a university veterinary hospital in South Korea (12.50%). In Europe, the highest frequency of CRGNFB was in veterinary hospitals in Italy (5.34%) (Table 3).
- Antimicrobial Susceptibility Evaluation
- Antimicrobial susceptibility in publications was evaluated using the agar diffusion (Kirby-Bauer) and minimum inhibitory concentration (MIC) techniques with standard and automated (Vitek bioMérieux, Marcy l'Étoile, Francia ; Wider (Francisco Soria Melguizo, SA, Madrid, Spain) and Sensitire (TREK Diagnostic Systems, Cleveland, OH, USA) broth microdilution and the epsilometry test (Epsilometer test (E test; AB Biodisk, Solna, Sweden). Two publications determined CR only with molecular techniques (Table S3). The MIC values of meropenem, imipenem, and ertapenem associated with CR isolates are presented in Table S3.
- Of 49 publications reporting CR phenotypes, 46 showed susceptibility data to antimicrobials other than carbapenems. CRE and CRGNFB exhibited resistance to penicillin; penicillin/β-lactamase inhibitors; cephamycins; first-, second-, third-, and fourth-generation cephalosporins; cephalosporins/β-lactamase inhibitors; monobactams; aminoglycosides; quinolones; sulfonamides; trimethoprim; tetracyclines; phenicols; nitrofurans; glycylcyclines; and polymyxins. Resistance to macrolides was evaluated and reported only in CRE. Susceptibility to phosphonic acids was evaluated, and resistance was reported in CRE and CRGNFB (Table S3). CRE and CRGNFB presented resistance to last-resort antimicrobials such as amikacin, colistin, fosfomycin, nitrofurantoin, and tigecycline in 14, 3, 8, 4, and 3 publications, respectively (Table S3).
- MDR isolates were reported in 20 articles in 4 CRE species (E. coli, K. pneumoniae, Salmonella enterica serovar Typhimurium, and E. cloacae) from companion animals and veterinary care surfaces. Four CRGNFB MDR species (P. aeruginosa, A. baumannii, Acinetobacter radioresistens, and Stenotrophomonas maltophilia) obtained from companion animals and domestic environments were reported in 7 articles (Table S3).
- The XDR phenotype was reported in 1 publication that highlighted 1 species of CRE (E. coli) from a canine, and 2 publications recording one species of CRGNFB (A. baumannii) obtained from dogs and cats. No isolates showed a PDR phenotype (Table S3).
- CR Genotypic Detection
- Genotypic CR was confirmed in publications employing 3 single or combined techniques: polymerase chain reaction, microarrays, and whole-genome sequencing (Table S3). In CRE, resistance was associated only with the production of carbapenemases. In companion animals, the “big five” carbapenemases were detected, with higher frequencies of OXA-48-like and NDM, and smaller proportions of KPC, IMP, and VIM. In veterinary environments, only OXA-48-like and NDM were detected. Conversely, only OXA-48 was found in commercial feed. None of the minor carbapenemases were identified in the CRE reported (Table 2).
- CR Acinetobacter spp. found in dogs and cats showed carbapenemase production as the only mechanism of resistance, with 2 of the “big five” (i.e., IMP and NDM), and the 3 important members of the genus (i.e., OXA-23-like, OXA-40-like, and OXA-58-like). CR P. aeruginosa was detected in dogs and home environments (sofa) and produced 2 of the 5 major carbapenemases (i.e., IMP and VIM). A CR mechanism different from carbapenemases was detected in a P. aeruginosa isolate from a canine. This mechanism involved the loss of the OprD outer membrane porin. CR S. maltophilia isolated from dogs and cats was associated with the production of L1, a species-intrinsic metallo-carbapenemase (Table 3).
- Geographical Distribution of CR Bacteria
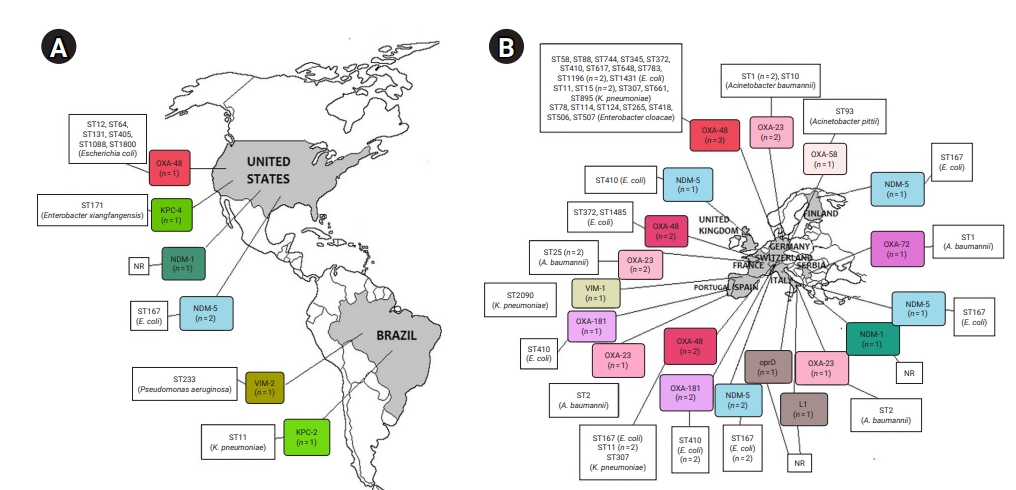
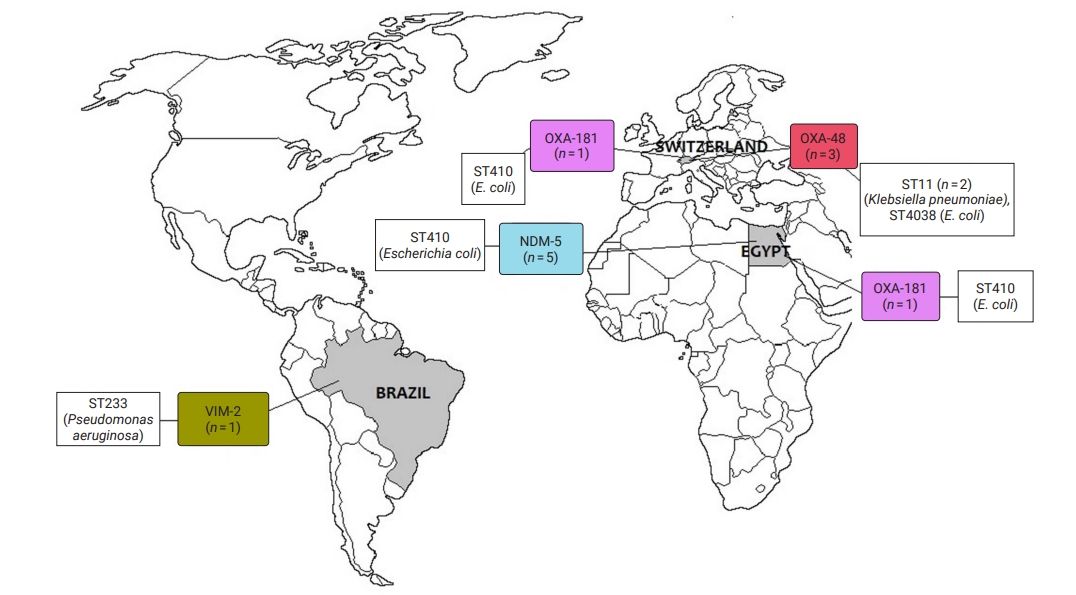
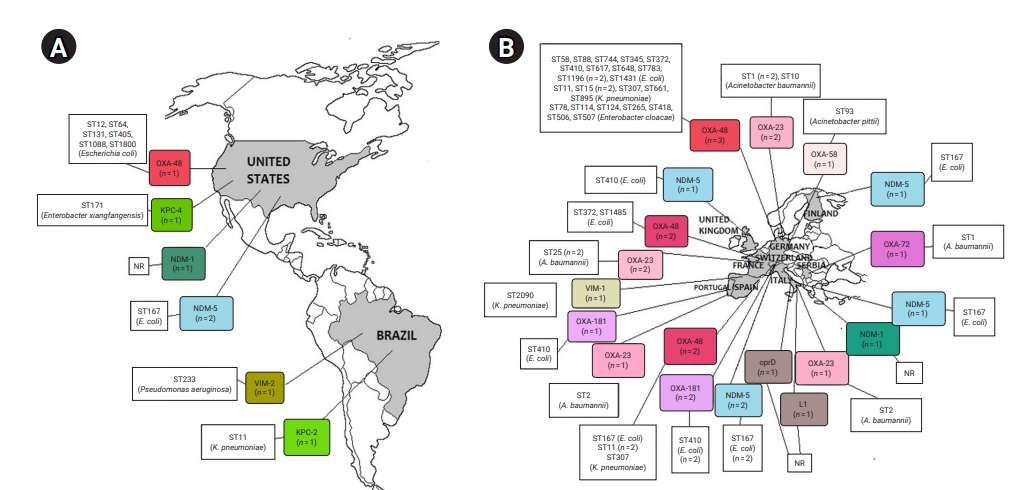
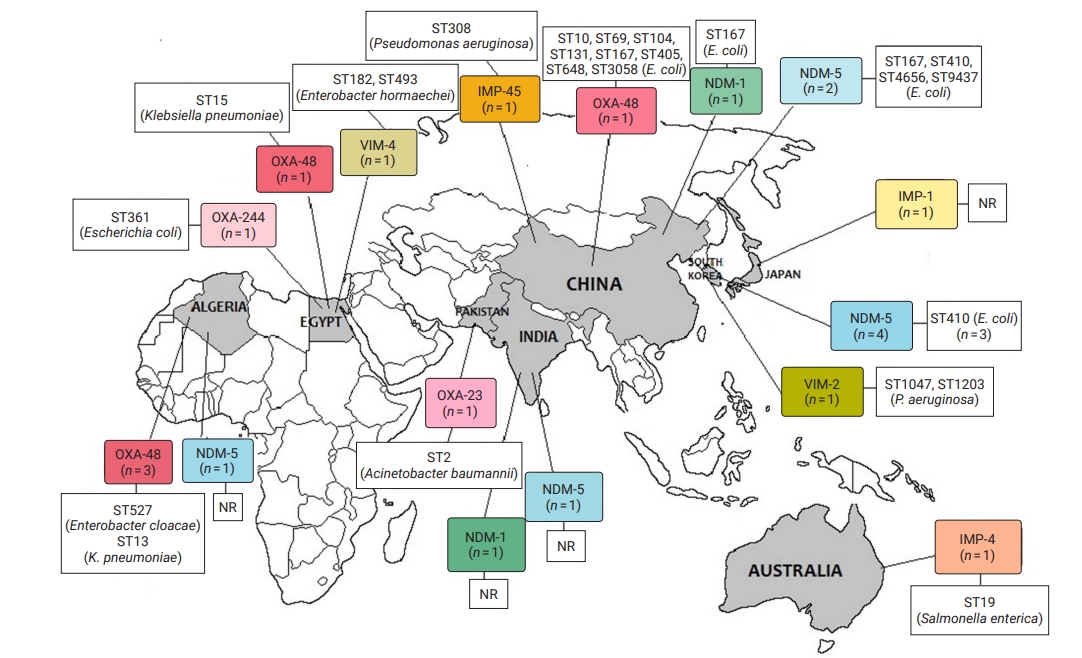
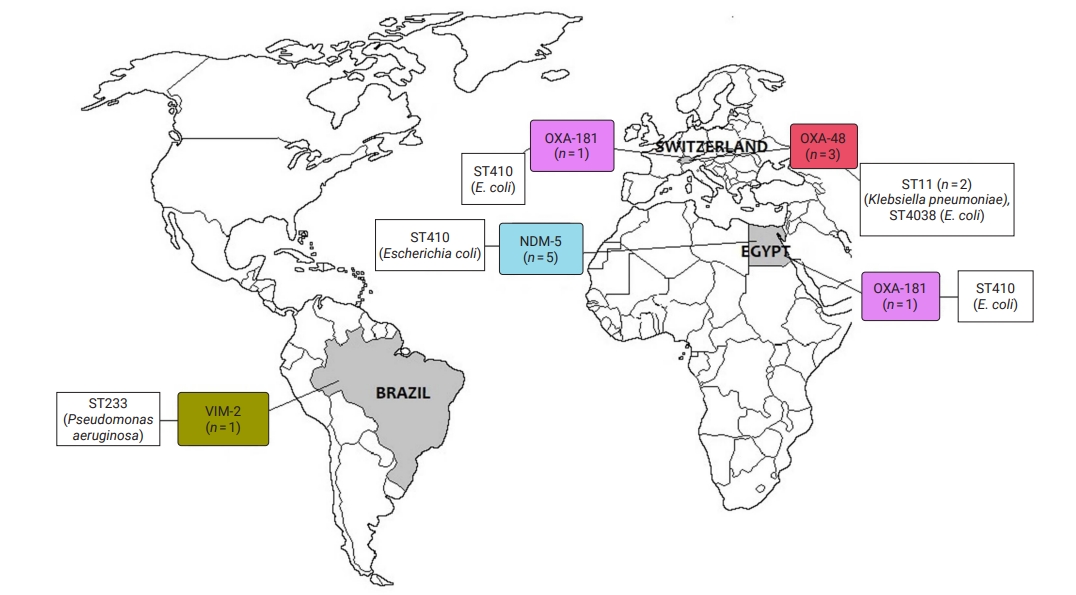
- CRE and CRGNFB acquired from companion animals were reported in 19 countries on 5 continents: 5 countries from Asia, 2 from the Americas, 9 from Europe, 2 from Africa, and 1 from Oceania (Figures 2 and 3). In companion animal-associated contexts, CRE and CRGNFB were reported in 3 countries on 3 continents: 1 in the Americas, 1 in Africa, and 1 in Europe (Figure 4).
- The blaNDM-5 gene was reported in plasmids only in CRE in Asia, the Americas, Europe, and Africa. The blaNDM-1 gene in CRE was detected in plasmids and chromosomes in Asia and the Americas; and in CRGNFB, without reporting its genomic location, but only in Europe. The blaOXA-48, blaOXA-181, and blaOXA-244 genes were found in plasmids only in CRE in Europe, the Americas, Africa, and Asia. The blaOXA-23, blaOXA-58, and blaOXA-72 genes located on plasmids and chromosomes were reported only in CRGNFB in Europe and Asia. The blaVIM-1 and blaVIM-4 genes on plasmids were registered in CRE in Europe and Africa, while the blaVIM-2 gene located on chromosomes and integrons was present in CRGNFB in the Americas and Asia. The blaKPC-2 and blaKPC-4 genes in plasmids were found exclusively in CRE in North and South America. The blaIMP-4 gene located on plasmids was reported only in CRE in Oceania, while the blaIMP-1 gene, with no registry of its genomic location, and blaIMP-45 on chromosomes were detected in CRGNFB in Asia (Table 1; Figures 2–4).
- Multilocus STs in CR Bacteria
- In publications that established the STs of CR bacteria identified by the molecular epidemiology technique of multilocus sequence typing, 55 STs were reported in CR isolates from companion animals, and 4 had descriptions of the contexts associated with their presence. Eight publications registered no STs. In CRE carrying blaNDM genes, 4 STs (ST4656, ST167, ST410, and ST9437) were reported on all continents except Oceania. CRE carrying blaVIM genes were clustered into 3 STs (ST2090, ST493, and ST182) in Africa and Europe, and CRGNFB into 3 STs (ST1047, ST1203, and ST233) in Asia and the Americas. Bacteria carrying blaIMP had the ST19 sequence in Oceania and the ST308 sequence in Asia in CRE and CRGNFB, respectively. Two STs (ST11 and ST171) were identified in blaKPC-producing CRE in the Americas (ST11, ST171). Bacteria carrying blaOXA had the widest variety of STs, with 39 in CRE in Asia, the Americas, Europe, and Africa and 5 in CRGNFB (ST1, ST10, ST2, ST25, and ST93) in Europe and Asia (Table 1; Figures 2–4).
- CR Isolations from Companion Animals Related to Humans and Other Animal Species
- Eleven studies that reported CR isolates in dogs and cats also registered other animal species, including pigs, birds (poultry, pet, and wild), cattle, sheep, goats, guinea pigs, rats, mice, rabbits, horses, fish, and flies. Seven publications reported CR in humans. Two were from hospital and community settings not related to companion animals, 2 from owners and employees in veterinary care settings, and 1 from backyard swine farm residents.
- In addition, 2 articles molecularly confirmed human-animal transmission from owners to dogs. CRE in Finland and CRGNFB in Brazil showed confirmed transmission with the same types of sequences and CR genes found in dogs. Both owners reported previous hospitalization, and 1 of them had also traveled internationally [34,54] (Table S4).
Results
- The current review compiled evidence on the presence and spread on 5 continents of Gram-negative bacteria categorized as critical in the WHO priority list of antibiotic-resistant bacteria for research and development of new antibiotics, such as CRE and CRGNFB isolated from domestic canines and felines, and on the contexts associated with their presence.
- Only 20% of the studies originated from surveillance programs, all in high-income and upper-middle-income countries. Since 2018, the United States has included dogs and cats in AMR surveillance programs [70,71]. In Europe, some countries included both species in their specific AMR control programs, and the European Union plans to launch the European Antimicrobial Resistance Surveillance Network in Veterinary Medicine (EARS-Vet), in which dogs and cats will be included in the scope of surveillance [72,73].
- However, the role of domestic canines and felines in CR bacterial transmission is often underestimated in local AMR containment programs [74,75]. Although in this study, the CR frequency in domestic canines and felines ranges from 0.4% to 6%, it is essential to integrate these animals into surveillance, control, and prevention strategies for CR, especially in low- and lower-middle-income countries where the problem may be underdiagnosed [76].
- Of particular concern are reports of VIM-2-producing P. aeruginosa [61] and an outbreak of OXA-181-producing E. coli [20] in veterinary hospitals in South Korea and Switzerland, with frequencies close to 12% and 20%, respectively. Although neither of those 2 reports described the administration of carbapenems, the use of meropenem in veterinary hospitals in South Korea [22,32,68] and the use of β-lactams and quinolones in the Swiss veterinary hospital have been registered, favoring the co-selection of CR isolates [34,59,77].
- Carbapenemase production was the most important mechanism of CR in Enterobacterales and GNFB of companion animals. The most frequent carbapenemase was OXA. Most OXA enzymes with carbapenemase activity have been identified in Acinetobacter spp. and P. aeruginosa [78]. The carbapenemases OXA-23-like (variant OXA-23), OXA-40-like (variant OXA-72), and OXA-58-like (variant OXA-58) were only identified in Acinetobacter spp. in companion animals from Europe and South Asia.
- However, the OXA-48-like enzyme in humans has been reported mainly in K. pneumoniae, E. coli, and E. cloacae [79], and it has been identified in pets, related environments, and food (variants OXA-48, OXA-181, and OXA-244). All were plasmid-encoded, confirming the risk posed by their rapid and easy transfer [79].
- The blaOXA-48 gene detected in the European countries in animal feed might be associated with the components of the feed formulation. blaOXA-48 has also been reported on Enterobacteriaceae obtained from poultry and swine carcasses in Europe and Asia [80] and drinking water systems of industrialized countries (United States) [81]. Conversely, the most likely source in this case, is human intervention during manufacturing and before packing [50], suggesting the relevance of humans in contaminating animal feed with CR organisms.
- The second most frequent enzyme found in this review was NDM in E. coli, E. cloacae, Citrobacter freundii, and A. radioresistens in companion animals in North America, Asia, Europe, and North African veterinary settings. NDM is considered endemic in the Balkan countries, the Middle East, and India, although it has spread worldwide, mainly through E. coli, K. pneumoniae, and Acinetobacter spp. [81−83]. E. coli ST101 and ST131 and K. pneumoniae ST11 and ST147 have been reported as epidemic clones responsible for its dissemination [81,83,84]. In E. coli, NDM was localized on plasmids. Nevertheless, E. coli presented ST167 and ST410 sequences, suggesting different dynamics in the circulation of NDM-producing E. coli in companion animals.
- Domestic canines and felines are not considered the main sources of CR acquisition for humans [81]. However, the transmission of CRE and CRGNFB from humans to dogs, from which the same microorganisms were isolated, has been reported in Finland [34] and Brazil [54], respectively. In both cases, the owners had a history of international travel or prolonged hospitalization, which are risk factors associated with CR acquisition [78,81].
- At the human-animal interface, the role of domestic canines and felines in CR dissemination in community settings should not be underestimated. In Brazil, the transmission of a CR bacterium between an owner and pet was confirmed, as well as its presence on shared household surfaces, such as the sofa [54]. The home can become a source of CR bacteria for companion animals, contributing to AMR spread in human environments. Canines, due to their generally more social behavior than felines, interact daily with other congeners and with people outside their family context, including at parks, daycare centers, shelters/kennels, and veterinary hospitals [85], favoring the spread/increase of the presence of CR bacteria in community settings.
- The presence of plasmid-borne IMP carbapenemase detected in the zoonotic bacterium Salmonella serovar Typhimurium in hospitalized cats in Australia [35] is of particular concern. Its presence could be attributed to human factors, considering that the circulation of the blaIMP gene has been demonstrated in Gram-negative bacteria in Australian human clinical settings and in migratory birds carrying CR Salmonella spp. acquired from human environments and genetically related to human isolates [86].
- Companion animals may also be involved in the spread of AMR in rural settings. A Chinese backyard pig farm reported CRE circulation in humans, birds, and flies, predominantly originating from canine gene complexes [19]. Human and animal populations that coexist in small-scale agricultural productions with insufficient biosecurity measures have been reported to be more vulnerable to acquiring CR [76]. In rural settings, the use of carbapenems may be lower due to the associated cost. However, using other antimicrobials that may favor co-selection and the occurrence of CR cannot be ruled out [34,59,77].
- The limitations of the current review include the fact that it was based only on studies published in electronic databases, and some valid reports of carbapenemases in companion animals in the gray literature may not have been identified unknown. Furthermore, low- and lower-middle-income countries may have been underrepresented due to the absence of publications in electronic databases. In addition, methods of isolation and detection of CR bacteria, phenotypic interpretation criteria, and the classification of resistant, intermediate, and susceptible isolates varied between the studies, and these factors may have influenced the collective results.
Discussion
- In conclusion, evidence of the presence of CRE and CRGNFB from companion animals and associated contexts in the 5 continents is compiled in this review. Domestic canines and felines are recognized as a possible source of dissemination and maintenance of carbapenemases for animals and humans. Thus, there is an urgent need for in-depth studies on the dynamics of CR circulation, including companion animals, under the concept of One Health in CR surveillance programs and plans for the containment of AMR, especially in low- and lower-middle-income countries, where the magnitude of the problem may be underestimated.
Conclusion
Supplementary Material
Table S1.
Table S2.
Table S3.
Table S4.
-
Ethics Approval
Not applicable.
-
Conflicts of Interest
The authors have no conflicts of interest to declare.
-
Funding
None.
-
Availability of Data
All data generated or analyzed in this study were included in this published article. More information can be requested from the corresponding author.
Article information
| Country | First author | Microorganism | CR mechanisma) | Genetic location | Study type |
|---|---|---|---|---|---|
| Companion animals | |||||
| China | Li et al. [19] | ENB | NDM-5 | Plasmid | Investigation (search) |
| Wang et al. [21] | ENB | NDM-5 | Plasmid | Investigation (search) | |
| Cui et al. [31] | ENB | NDM-1 | Plasmid | Surveillance | |
| Liu et al. [39] | ENB | OXA-48 | NR | Investigation (search) | |
| Wang et al. [53] | GNFB | IMP-45 | Chromosome | Surveillance | |
| South Korea | Hong et al. [22] | ENB | NDM-5 | Plasmid | Investigation (report) |
| Hong et al. [32] | ENB | NDM-5 | Plasmid | Surveillance | |
| Hong et al. [45] | ENB | NDM-5 | NR | Surveillance | |
| Oh et al. [68] | ENB | NDM-5 | Plasmid | Monitoring | |
| Hyun et al. [61] | GNFB | VIM-2 | Integron class I | Investigation (search) | |
| India | Pruthvishree et al. [46] | ENB | NDM-1 | NR | Investigation (report) |
| Bandyopadhyay et al. [67] | ENB | NDM-5 | Plasmid | Investigation (search) | |
| Japan | Kimura et al. [65] | GNFB | IMP-1 | NR | Surveillance |
| Pakistan | Taj et al. [66] | GNFB | OXA-23 | NR | Investigation (report) |
| Brazil | Sellera et al. [42] | ENB | KPC-2 | Plasmid | Surveillance |
| Fernandes et al. [54] | GNFB | VIM-2 | Chromosome | Investigation (report and search) | |
| United States | Liu et al. [36] | ENB | OXA-48 | Plasmid | Investigation (search) |
| Daniels et al. [41] | ENB | KPC-4 | Plasmid | Surveillance | |
| Shaheen et al. [47] | ENB | NDM-1 | Plasmid and chromosome | Investigation (search) | |
| Tyson et al. [60] | ENB | NDM-5 | Plasmid | Surveillance | |
| Cole et al. [69] | ENB | NDM-5 | Plasmid | Investigation (search) | |
| Germany | Pulss et al. [25] | ENB | OXA-48 | Plasmid | Investigation (search) |
| Stolle et al. [29] | ENB | OXA-48 | Plasmid | Investigation (search) | |
| Schmiedel et al. [37] | ENB | OXA-48 | NR | Investigation (search) | |
| Ewers et al. [55] | GNFB | OXA-23 | Plasmid | Investigation (search) | |
| Ewers et al. [57] | GNFB | OXA-23 | Plasmid | Investigation (report) | |
| Klotz et al. [63] | GNFB | OXA-58 | Plasmid | Investigation (search) | |
| Spain | Gonzalez-Torralba et al. [33] | ENB | VIM-1 | Plasmid | Investigation (search) |
| Finland | Gronthal et al. [34] | ENB | NDM-5 | Plasmid | Investigation (report and search) |
| France | Valat et al. [23] | ENB | OXA-48 | Plasmid | Investigation (search) |
| Melo et al. [48] | ENB | OXA-48 | Plasmid | Investigation (search) | |
| Herivaux et al. [56] | GNFB | OXA-23 | NR | Investigation (search) | |
| Lupo et al. [64] | GNFB | OXA-23 | Chromosome | Surveillance | |
| Italy | Alba et al. [49] | ENB | NDM-5 | Plasmid | Investigation (report) |
| Gentilini et al. [59] | GNFB | NDM-1, OXA-23, L1b), LP | NR | Investigation (search) | |
| Portugal | Brilhante et al. [26] | ENB | OXA-181 | Plasmid | Investigation (search) |
| Pomba et al. [58] | GNFB | OXA-23 | Chromosome | Investigation (report) | |
| United Kingdom | Reynolds et al. [24] | ENB | NDM-5 | Plasmid | Surveillance |
| Serbia | Misic et al. [62] | GNFB | OXA-72 | Plasmid | Investigation (report) |
| Switzerland | Nigg et al. [20] | ENB | OXA-181 | Plasmid | Investigation (search) |
| Brilhante et al. [38] | ENB | OXA-48 | Plasmid | Investigation (report) | |
| Dazio et al. [40] | ENB | NDM-5, OXA-48, OXA-181 | NR | Investigation (search) | |
| Peterhans et al. [43] | ENB | NDM- 5 | Plasmid | Investigation (NR) | |
| Algeria | Yousfi et al. [27] | ENB | OXA-48, NDM-5 | NR | Investigation (search) |
| Yousfi et al. [28] | ENB | OXA-48 | Plasmid | Investigation (search) | |
| Mairi et al. [30] | ENB | OXA-48 | Plasmid | Investigation (search) | |
| Egypt | Khalifa et al. [44] | ENB | VIM-4, OXA-48, OXA-244 | Plasmid | Investigation (search) |
| Australia | Abraham et al. [35] | ENB | IMP-4 | Plasmid | Investigation (report) |
| Context associated with companion animalsc) | |||||
| Brazil | Fernandes et al. [54] | GNFB | VIM-2 | Chromosome | Investigation (report and search) |
| Egypt | Ramadan et al. [51] | ENB | NDM-5, OXA-181 | Plasmid | Investigation (search) |
| Switzerland | Brilhante et al. [38] | ENB | OXA-48 | Plasmid | Investigation (report) |
| Seiffert et al. [50] | ENB | OXA-48 | Plasmid | Investigation (search) | |
| Schmidt et al. [52] | ENB | OXA-48, OXA-181 | NR | Investigation (search) |
CR, carbapenem resistant; ENB, enterobacteria; NDM, New Delhi metallo-β-lactamase; OXA, oxacillin; NR, not reported; GNFB, glucose non-fermenting bacilli; IMP, active-on-imipenem; VIM, Verona integron-encoded metallo-β-lactamase; KPC, Klebsiella pneumoniae carbapenemase; LP, loss of porine.
a) Carbapenemase production or loss of porins.
b) Intrinsic metallo-β-lactamase L1 of the species Stenotrophomonas maltophilia encoded by chromosomes.
c) Veterinary medical care surfaces, household surfaces, and companion animal food.
| Study |
Sample origin |
Antimicrobial use |
Carbapenem resistance |
||||
|---|---|---|---|---|---|---|---|
| Animal | n | Specimen | Carbapenem | Others | Bacterial species | CR origina) | |
| Li et al. [19] | Dogs | 2 | Stool | NR | NR | Escherichia coli | NDM-5 |
| Nigg et al. [20] | Dogs, cats | 21 | Rectal swab | NO | PEN, BET/INHIB, QUIN, NIT, CEP, TET, SUL | E. coli | OXA-181 (OXA-48-like) |
| Wang et al. [21] | Dogs, cats | 6 | Stool, urine | NR | NR | E. coli, Enterobacter cloacae, Citrobacter freundii | NDM-5 |
| Hong et al. [22] | Dogs, cats | 2 | Rectal swab | MERO | QUIN, CEP, TET | E. coli | NDM-5 |
| Valat et al. [23] | Dogs | 1 | NR | NR | NR | E. coli | OXA-48 |
| Reynolds et al. [24] | Dogs | 1 | Tissuesb) | NO | BET/INHIB, CEP, TET, QUIN | E. coli | NDM-5 |
| Pulss et al. [25] | Dogs, cats | 130 | CVADsc), urine, other fluidsd), tissuesb) | NO | CEP 2°, 3°, PEN, QUIN, BET/INHB | Klebsiella pneumoniae/Klebsiella oxytoca, E. cloacae, E. coli | OXA-48 |
| Brilhante et al. [26] | Dogs | 1 | Tissuesb) | NR | NR | E. coli | OXA-181 (OXA-48-like) |
| Yousfi et al. [27] | Dogs, cats | 5 | Rectal swab | NR | NR | E. coli | OXA-48, NDM-5 |
| Yousfi et al. [28] | Dogs, cats | 6 | Rectal swab | NR | NR | E. coli, K. pneumoniae, E. cloacae | OXA-48 |
| Stolle et al. [29] | Dogs | 6 | CVADsc), tissuesb), stool, urine, other fluidsd) | NO | PEN, BET/INHIB, QUIN, CEP, 3° TET | K. pneumoniae, E. coli | OXA-48 |
| Mairi et al. [30] | Dogs | 1 | Rectal swab | NR | NR | K. pneumoniae | OXA-48 |
| Cui et al. [31] | Dogs | 1 | Anal swab | NR | NR | E. coli | NDM-1 |
| Hong et al. [32] | Dogs | 4 | Rectal swab | MERO | CEP 1°, QUIN, BET/INHIB, NIT | E. coli | NDM-5 |
| Gonzalez-Torralba et al.[33] | Dogs | 1 | Rectal swab | NO | NO | K. pneumoniae | VIM-1 |
| Gronthal et al. [34] | Dogs | 2 | Ear swab | NO | QUIN, CEP 1°, BET/INHIB, LIN | E. coli | NDM-5 |
| Abraham et al. [35] | Cats | 4 | Stool | NO | TET | Salmonella enterica serovar Typhimurium | IMP-4 |
| Liu et al. [36] | Dogs, cats | NR | Urine, tissuesb) | NR | NR | E. coli | OXA-48 |
| Schmiedel et al. [37] | Dogs, cats | NR | NR | NR | NR | K. pneumoniae, E. cloacae | OXA-48 |
| Brilhante et al. [38] | Dogs, cats | 10 | Urine, other fluidsd), tissuesb) | NR | NR | K. pneumoniae | OXA-48 |
| Liu et al. [39] | Dogs | NR | Urine, tissuesb) | NR | NR | E. coli | OXA-48 |
| Dazio et al. [40] | Dogs, cats | 25 | Rectal swab | NO | NR | E. coli | NDM-5, OXA-48, OXA-181 (OXA-48-like) |
| K. pneumoniae | OXA-48 | ||||||
| Daniels et al. [41] | Dogs | 2 | Urine, tissuesb) | NO | TET | Enterobacter. hormaechei subsp. xiangfangensis | KPC-4 |
| Sellera et al. [42] | Dogs | 1 | Urine | NR | NR | K. pneumoniae | KPC-2 |
| Peterhans et al. [43] | Dogs | 1 | Tissuesb) | NR | NR | E. coli | NDM-5 |
| Khalifa et al. [44] | Dogs, cats | 7 | Nasal swab, eye swab | NR | NR | E. hormaechei subsp. xiangfangensis | VIM-4 |
| K. pneumoniae | OXA-48 | ||||||
| E. coli | OXA-244 (OXA-48-like) | ||||||
| Hong et al. [45] | Dogs | 4 | Rectal swab | NR | NR | E. coli | NDM-5 |
| Pruthvishree et al. [46] | Dogs | 1 | Other fluidsd) | NR | NR | E. coli | NDM-1 |
| Shaheen et al. [47] | Dogs, cats | 6 | Urine, tissuesb) | NR | NR | E. coli | NDM-1 |
| Melo et al. [48] | Dogs | 1 | Rectal swab | NO | BET/INHIB | E. coli | OXA-48 |
| Alba et al. [49] | Dogs | 1 | Urine | NR | NR | E. coli | NDM-5 |
| Wang et al. [53] | Dogs | 1 | Anal swab | NR | NR | Pseudomonas aeruginosa | IMP-45 |
| Fernandes et al. [54] | Dogs | 1 | Oral swab, rectal swab, other fluidsd) | NR | NR | P. aeruginosa | VIM-2 |
| Ewers et al. [55] | Dogs, cats | 3 | Vaginal swab, urine, other fluidsd) | NR | NR | Acinetobacter baumannii | OXA-23 |
| Herivaux et al. [56] | Dogs | 2 | Oral swab, rectal swab | NO | NO | A. baumannii | OXA-23 |
| Ewers et al. [57] | Cats | 1 | Urine | NR | NR | A. baumannii | OXA-23 |
| Pomba et al. [58] | Cats | 1 | Urine | NO | BET/INHB | A. baumannii | OXA-23 |
| Gentilini et al. [59] | Dogs, cats | 11 | Rectal swab | NO | BET/INHIB, TET, QUIN, NIT | A. radioresistens | NDM-1 |
| A. baumannii | OXA-23 | ||||||
| P. aeruginosa | Loss of porine) | ||||||
| Stenotrophomonas maltophilia | L1f) | ||||||
| Tyson et al. [60] | Dogs | 1 | Other fluidsd) | NR | NR | E. coli | NDM-5 |
| Hyun et al. [61] | Dogs | 10 | Other fluidsd), tissuesb) | NR | NR | P. aeruginosa | VIM- 2 |
| Misic et al. [62] | Dogs | 1 | Urine | NR | NR | A. baumannii | OXA-72 (OXA-40-like) |
| Klotz et al. [63] | Dogs, cats | 4 | Nasal swab, other fluidsd), tissuesb) | NR | NR | Acinetobacter pittii | OXA-58 (OXA-58-like) |
| Lupo et al. [64] | Dogs, cats | 7 | NR | NR | NR | A. baumannii | OXA-23 |
| Kimura et al. [65] | Dogs, cats | 2 | Urine, other fluidsd) | NO | PHOS | A. radioresistens | IMP-1 |
| Taj et al. [66] | Cats | 1 | Urine | NO | BET/ INHIB, QUIN | A. baumannii | OXA-23 |
| Bandyopadhyay et al. [67] | Dogs | 16 | Rectal swab, vaginal swab, tissuesb) | NR | NR | E. coli | NDM-5 |
| Oh et al. [68] | Dogs | 4 | Stool, nasal swab, urine | MERO | BET/ INHIB, TET | E. coli | NDM-5 |
| Cole et al. [69] | Dogs, cats | 6 | Urine, other fluidsd), tissuesb) | NO | PEN, CEP, AMN, NIT, BET/ INHIB | E. coli | NDM-5 |
| Brilhante et al. [38] | Veterinary surfaces | NA | Environmental swabs | NA | K. pneumoniae | OXA-48 | |
| Seiffert et al. [50] | Pet foodg) | NA | Pet food packages | NA | Enterobacterales (undetermined species) | OXA-48 | |
| Ramadan et al. [51] | Veterinary surfaces | NA | Environmental swabs | NA | E. coli | NDM-5, OXA-181 (OXA-48-like) | |
| Schmidt et al. [52] | Veterinary surfaces | NA | Environmental swabs | NA | E. coli | OXA-48, OXA-181 (OXA-48-like) | |
| K. pneumoniae | OXA-48 | ||||||
| E. cloacae | OXA-48 | ||||||
| Fernandes et al. [54] | Household surfacesh) | NA | Environmental swabs | NA | P. aeruginosa | VIM-2 | |
CR, carbapenem resistant; NR, not reported; NDM, New Delhi metallo-β-lactamase; NO, non-use; PEN, penicillins; BET/INHIB, beta-lactams/beta-lactam inhibitors; QUIN, quinolones; NIT, nitroimidazoles; CEP, cephalosporins; TET, tetracyclines; SUL, sulfonamides; OXA, oxacillin; MERO, meropenem; CVADs, central venous access devices; VIM, Verona integron-encoded metallo-β-lactamase; LIN, lincosamides; IMP, active-on-imipenem; KPC, Klebsiella pneumoniae carbapenemase; PHOS, phosphonic acids; NA, not applicable; AMN, aminoglycosides.
a) Referred to the mechanism that confers resistance to the microorganism: production of carbapenemases or loss of the bacterial target.
b) Included wound tissues, nasal structure, skin, abdominal cavity, anal sacs, intestine, and lung.
c) Included central venous catheters.
d) Included bronchoalveolar and tracheobronchial lavage, scrotal fluid, pus, bile, ear discharge, and eye discharge.
e) The authors determined that the loss of the oprD porin was caused by different mutations within the gene that caused a premature stop codon because of a large insertion, a frameshift, or a nonsense mutation.
f) Chromosome-encoded intrinsic metallo-β-lactamase L1 of S. maltophilia species.
g) Mixed wet pet food with different flavors.
h) Home surfaces (sofa, balcony, water cooler).
| First author | Country | Isolation place |
Sampled animals (n) |
Animals with CR microorganisms (n) |
Frequency of animals with CR (%) |
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Dogs | Cats | Total | Dogs | Cats | Total | Dogs | Cats | Total | |||
| Li et al. [19] | China | Households with backyard pig farms | 92 | 19 | 111 | 2 | 0 | 2 | 2.17 | 0.00 | 1.80 |
| Nigg et al. [20] | Switzerland | University veterinary hospital | 76 | 21 | 97 | 17 | 4 | 21 | 22.37 | 19.05 | 21.65 |
| Reynolds et al. [24] | United Kingdom | Veterinary clinics | 158 | 27 | 185 | 1 | 0 | 1 | 0.63 | 0.00 | 0.54 |
| Pulss et al. [25] | Germany | Veterinary microbiology laboratory | 3,375 | 932 | 4,307 | 117 | 13 | 130 | 3.47 | 1.39 | 3.02 |
| Brilhante et al. [26] | Portugal | Homes and university veterinary hospital | 71 | 27 | 98 | 1 | 0 | 1 | 1.41 | 0.00 | 1.02 |
| Yousfi et al. [27] | Algeria | Local veterinary office | 116 | 84 | 200 | 3 | 2 | 5 | 2.59 | 2.38 | 2.50 |
| Yousfi et al. [28] | Algeria | Local veterinary office and homes | 265 | 49 | 314 | 4 | 2 | 6 | 1.51 | 4.08 | 1.91 |
| Mairi et al. [30] | Algeria | NR | 75 | 37 | 112 | 1 | 0 | 1 | 1.33 | 0.00 | 0.89 |
| Cui et al. [31] | China | University veterinary hospital | NR | NR | 226 | 1 | 0 | 1 | NA | NA | 0.44 |
| Hong et al. [32] | South Korea | Veterinary clinics | 353 | 0 | 353 | 4 | 0 | 4 | 1.13 | 0.00 | 1.13 |
| Gonzalez-Torralba et al. [33] | Spain | Companion animal shelter | 160 | 0 | 160 | 1 | 0 | 1 | 0.63 | 0.00 | 0.63 |
| Khalifa et al. [44] | Egypt | NR | NR | NR | 1,348 | 3 | 4 | 7 | NA | NA | 0.52 |
| Hong et al. [45] | South Korea | Veterinary hospitals | 315 | 74 | 389 | 4 | 0 | 4 | 1.27 | 0.00 | 1.03 |
| Melo et al. [48] | France | University veterinary hospital | 166 | 227 | 393 | 1 | 0 | 1 | 0.60 | 0.00 | 0.25 |
| Bandyopadhyay et al. [67] | India | Veterinary clinics and university veterinary hospital | 237 | 0 | 237 | 16 | 0 | 16 | 6.75 | 0.00 | 6.75 |
| Herivaux et al. [56] | France | University veterinary hospital | 104 | 46 | 150 | 2 | 0 | 2 | 1.92 | 0.00 | 1.33 |
| Gentilini et al. [59] | Italy | Veterinary hospitals and homes | 134 | 72 | 206 | 8 | 3 | 11 | 5.97 | 4.17 | 5.34 |
| Hyun et al. [61] | South Korea | Veterinary medical teaching hospital | 80 | 0 | 80 | 10 | 0 | 10 | 12.5 | 0.00 | 12.50 |
| Klotz et al. [63] | Germany | Veterinary clinics | 110 | 48 | 158 | 2 | 2 | 4 | 1.82 | 4.17 | 2.53 |
- 1. Antimicrobial Resistance Collaborators. Global burden of bacterial antimicrobial resistance in 2019: a systematic analysis. Lancet 2022;399:629−55.PubMedPMC
- 2. Prestinaci F, Pezzotti P, Pantosti A. Antimicrobial resistance: a global multifaceted phenomenon. Pathog Glob Health 2015;109:309−18.ArticlePubMedPMC
- 3. Centers for Disease Control and Prevention (CDC). Antibiotic resistance threats in the United States [Internet]. Atlanta: CDC; 2019 [cited 2021 Jun 15]. Available from: https://www.cdc.gov/drugresistance/pdf/threats-report/2019-ar-threats-report-508.pdf.
- 4. Lemos EV, de la Hoz FP, Alvis N, et al. Impact of carbapenem resistance on clinical and economic outcomes among patients with Acinetobacter baumannii infection in Colombia. Clin Microbiol Infect 2014;20:174−80.ArticlePubMed
- 5. Gottig S, Gruber TM, Higgins PG, et al. Detection of pan drug-resistant Acinetobacter baumannii in Germany. J Antimicrob Chemother 2014;69:2578−9.ArticlePubMed
- 6. Bush K. The importance of β-lactamases to the development of new β-lactams. Edited by Mayers DL, Sobel JD, Ouellette M, et al.: Antimicrobial drug resistance. Cham: Springer; 2017. pp 165−74.
- 7. Lv L, Zeng Z, Song Q, et al. Emergence of XDR Escherichia coli carrying both blaNDM and mcr-1 genes in chickens at slaughter and the characterization of two novel blaNDM-bearing plasmids. J Antimicrob Chemother 2018;73:2261−3.ArticlePubMed
- 8. Bonnin RA, Jousset AB, Emeraud C, et al. Genetic diversity, biochemical properties, and detection methods of minor carbapenemases in Enterobacterales. Front Med (Lausanne) 2021;7:616490. ArticlePubMedPMC
- 9. Ambler RP. The structure of beta-lactamases. Philos Trans R Soc Lond B Biol Sci 1980;289:321−31.PubMed
- 10. Bush K, Jacoby GA. Updated functional classification of beta-lactamases. Antimicrob Agents Chemother 2010;54:969−76.ArticlePubMedPDF
- 11. Tacconelli E, Carrara E, Savoldi A, et al. Discovery, research, and development of new antibiotics: the WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect Dis 2018;18:318−27.PubMed
- 12. Hamidian M, Nigro SJ. Emergence, molecular mechanisms and global spread of carbapenem-resistant Acinetobacter baumannii. Microb Genom 2019;5:e000306.ArticlePubMedPMC
- 13. Mentasti M, Prime K, Sands K, et al. Rapid detection of OXA-23-like, OXA-24-like, and OXA-58-like carbapenemases from Acinetobacter species by real-time PCR. J Hosp Infect 2020;105:741−6.ArticlePubMed
- 14. Committee for Medicinal Products for Veterinary Use (CVMP), Committee for Medicinal Products for Human Use (CHMP). Categorisation of antibiotics in the European Union [Internet]. Amsterdam: European Medicines Agency; 2019 [cited 2021 Jun 15]. Available from: https://www.ema.europa.eu/en/documents/report/categorisation-antibiotics-european-union-answer-request-european-commission-updating-scientific_en.pdf.
- 15. Abraham S, Wong HS, Turnidge J, et al. Carbapenemase-producing bacteria in companion animals: a public health concern on the horizon. J Antimicrob Chemother 2014;69:1155−7.ArticlePubMed
- 16. Pomba C, Rantala M, Greko C, et al. Public health risk of antimicrobial resistance transfer from companion animals. J Antimicrob Chemother 2017;72:957−68.ArticlePubMed
- 17. One Health High-Level Expert Panel (OHHLEP), Adisasmito WB, Almuhairi S, et al. One Health: a new definition for a sustainable and healthy future. PLoS Pathog 2022;18:e1010537.ArticlePubMedPMC
- 18. Collignon PJ, McEwen SA. One Health: its importance in helping to better control antimicrobial resistance. Trop Med Infect Dis 2019;4:22. ArticlePubMedPMC
- 19. Li J, Bi Z, Ma S, et al. Inter-host transmission of carbapenemase-producing Escherichia coli among humans and backyard animals. Environ Health Perspect 2019;127:107009. ArticlePubMedPMC
- 20. Nigg A, Brilhante M, Dazio V, et al. Shedding of OXA-181 carbapenemase-producing Escherichia coli from companion animals after hospitalisation in Switzerland: an outbreak in 2018. Euro Surveill 2019;24:1900071. ArticlePubMedPMC
- 21. Wang J, Xia YB, Huang XY, et al. Emergence of blaNDM-5 in Enterobacteriaceae isolates from companion animals in Guangzhou, China. Microb Drug Resist 2021;27:809−15.ArticlePubMed
- 22. Hong JS, Song W, Jeong SH. Molecular characteristics of NDM-5-producing Escherichia coli from a cat and a dog in South Korea. Microb Drug Resist 2020;26:1005−8.ArticlePubMed
- 23. Valat C, Drapeau A, Beurlet S, et al. Pathogenic Escherichia coli in dogs reveals the predominance of ST372 and the human-associated ST73 extra-intestinal lineages. Front Microbiol 2020;11:580. ArticlePubMedPMC
- 24. Reynolds ME, Phan HT, George S, et al. Occurrence and characterization of Escherichia coli ST410 co-harbouring blaNDM-5, blaCMY-42 and blaTEM-190 in a dog from the UK. J Antimicrob Chemother 2019;74:1207−11.ArticlePubMed
- 25. Pulss S, Stolle I, Stamm I, et al. Multispecies and clonal dissemination of OXA-48 carbapenemase in Enterobacteriaceae from companion animals in Germany, 2009-2016. Front Microbiol 2018;9:1265. ArticlePubMedPMC
- 26. Brilhante M, Menezes J, Belas A, et al. OXA-181-producing extraintestinal pathogenic Escherichia coli sequence type 410 isolated from a dog in Portugal. Antimicrob Agents Chemother 2020;64:e02298−19.ArticlePubMedPMCPDF
- 27. Yousfi M, Touati A, Mairi A, et al. Emergence of carbapenemase-producing Escherichia coli isolated from companion animals in Algeria. Microb Drug Resist 2016;22:342−6.ArticlePubMed
- 28. Yousfi M, Touati A, Muggeo A, et al. Clonal dissemination of OXA-48-producing Enterobacter cloacae isolates from companion animals in Algeria. J Glob Antimicrob Resist 2018;12:187−91.ArticlePubMed
- 29. Stolle I, Prenger-Berninghoff E, Stamm I, et al. Emergence of OXA-48 carbapenemase-producing Escherichia coli and Klebsiella pneumoniae in dogs. J Antimicrob Chemother 2013;68:2802−8.ArticlePubMed
- 30. Mairi A, Pantel A, Ousalem F, et al. OXA-48-producing Enterobacterales in different ecological niches in Algeria: clonal expansion, plasmid characteristics and virulence traits. J Antimicrob Chemother 2019;74:1848−55.ArticlePubMedPDF
- 31. Cui L, Lei L, Lv Y, et al. blaNDM-1-producing multidrug-resistant Escherichia coli isolated from a companion dog in China. J Glob Antimicrob Resist 2018;13:24−7.ArticlePubMed
- 32. Hong JS, Song W, Park HM, et al. First detection of New Delhi Metallo-β-lactamase-5-producing Escherichia coli from companion animals in Korea. Microb Drug Resist 2019;25:344−9.ArticlePubMed
- 33. Gonzalez-Torralba A, Oteo J, Asenjo A, et al. Survey of carbapenemase-producing Enterobacteriaceae in companion dogs in Madrid, Spain. Antimicrob Agents Chemother 2016;60:2499−501.ArticlePubMedPMCPDF
- 34. Gronthal T, Osterblad M, Eklund M, et al. Sharing more than friendship: transmission of NDM-5 ST167 and CTX-M-9 ST69 Escherichia coli between dogs and humans in a family, Finland, 2015. Euro Surveill 2018;23:1700497. PubMedPMC
- 35. Abraham S, O’Dea M, Trott DJ, et al. Isolation and plasmid characterization of carbapenemase (IMP-4) producing Salmonella enterica Typhimurium from cats. Sci Rep 2016;6:35527. ArticlePubMedPMCPDF
- 36. Liu X, Thungrat K, Boothe DM. Occurrence of OXA-48 carbapenemase and other β-lactamase genes in ESBL-producing multidrug resistant Escherichia coli from dogs and cats in the United States, 2009-2013. Front Microbiol 2016;7:1057. ArticlePubMedPMC
- 37. Schmiedel J, Falgenhauer L, Domann E, et al. Multiresistant extended-spectrum β-lactamase-producing Enterobacteriaceae from humans, companion animals and horses in central Hesse, Germany. BMC Microbiol 2014;14:187. ArticlePubMedPMCPDF
- 38. Brilhante M, Gobeli Brawand S, Endimiani A, et al. Two high-risk clones of carbapenemase-producing Klebsiella pneumoniae that cause infections in pets and are present in the environment of a veterinary referral hospital. J Antimicrob Chemother 2021;76:1140−9.ArticlePubMedPDF
- 39. Liu X, Liu H, Li Y, et al. High prevalence of β-lactamase and plasmid-mediated quinolone resistance genes in extended-spectrum cephalosporin-resistant Escherichia coli from dogs in Shaanxi, China. Front Microbiol 2016;7:1843. ArticlePubMedPMC
- 40. Dazio V, Nigg A, Schmidt JS, et al. Acquisition and carriage of multidrug-resistant organisms in dogs and cats presented to small animal practices and clinics in Switzerland. J Vet Intern Med 2021;35:970−9.ArticlePubMedPMCPDF
- 41. Daniels JB, Chen L, Grooters SV, et al. Enterobacter cloacae complex sequence type 171 isolates expressing KPC-4 carbapenemase recovered from canine patients in Ohio. Antimicrob Agents Chemother 2018;62:e01161−18.ArticlePMCPDF
- 42. Sellera FP, Fuga B, Fontana H, et al. Detection of IncN-pST15 one-health plasmid harbouring blaKPC-2 in a hypermucoviscous Klebsiella pneumoniae CG258 isolated from an infected dog, Brazil. Transbound Emerg Dis 2021;68:3083−8.ArticlePubMedPMCPDF
- 43. Peterhans S, Stevens MJA, Nuesch-Inderbinen M, et al. First report of a blaNDM-5-harbouring Escherichia coli ST167 isolated from a wound infection in a dog in Switzerland. J Glob Antimicrob Resist 2018;15:226−7.ArticlePubMed
- 44. Khalifa HO, Oreiby AF, Abd El-Hafeez AA, et al. First report of multidrug-resistant carbapenemase-producing bacteria coharboring mcr-9 associated with respiratory disease complex in pets: potential of animal-human transmission. Antimicrob Agents Chemother 2020;65:e01890−20.ArticlePubMedPMCPDF
- 45. Hong JS, Song W, Park HM, et al. Clonal spread of extended-spectrum cephalosporin-resistant Enterobacteriaceae between companion animals and humans in South Korea. Front Microbiol 2019;10:1371. ArticlePubMedPMC
- 46. Pruthvishree BS, Vinodh Kumar OR, Sivakumar M, et al. Molecular characterization of extensively drug resistant (XDR), extended spectrum beta-lactamases (ESBL) and New Delhi Metallo beta-lactamase-1 (blaNDM1) producing Escherichia coli isolated from a male dog-a case report. Vet Arh 2018;88:139−48.Article
- 47. Shaheen BW, Nayak R, Boothe DM. Emergence of a New Delhi metallo-β-lactamase (NDM-1)-encoding gene in clinical Escherichia coli isolates recovered from companion animals in the United States. Antimicrob Agents Chemother 2013;57:2902−3.ArticlePubMedPMCPDF
- 48. Melo LC, Boisson MN, Saras E, et al. OXA-48-producing ST372 Escherichia coli in a French dog. J Antimicrob Chemother 2017;72:1256−8.ArticlePubMed
- 49. Alba P, Taddei R, Cordaro G, et al. Carbapenemase IncF-borne blaNDM-5 gene in the E. coli ST167 high-risk clone from canine clinical infection, Italy. Vet Microbiol 2021;256:109045. ArticlePubMed
- 50. Seiffert SN, Carattoli A, Tinguely R, et al. High prevalence of extended-spectrum β-lactamase, plasmid-mediated AmpC, and carbapenemase genes in pet food. Antimicrob Agents Chemother 2014;58:6320−3.ArticlePubMedPMCPDF
- 51. Ramadan H, Gupta SK, Sharma P, et al. Circulation of emerging NDM-5-producing Escherichia coli among humans and dogs in Egypt. Zoonoses Public Health 2020;67:324−9.ArticlePubMedPDF
- 52. Schmidt JS, Kuster SP, Nigg A, et al. Poor infection prevention and control standards are associated with environmental contamination with carbapenemase-producing Enterobacterales and other multidrug-resistant bacteria in Swiss companion animal clinics. Antimicrob Resist Infect Control 2020;9:93. ArticlePubMedPMCPDF
- 53. Wang Y, Wang X, Schwarz S, et al. IMP-45-producing multidrug-resistant Pseudomonas aeruginosa of canine origin. J Antimicrob Chemother 2014;69:2579−81.ArticlePubMed
- 54. Fernandes MR, Sellera FP, Moura Q, et al. Zooanthroponotic transmission of drug-resistant Pseudomonas aeruginosa, Brazil. Emerg Infect Dis 2018;24:1160−2.ArticlePubMedPMC
- 55. Ewers C, Klotz P, Leidner U, et al. OXA-23 and ISAba1-OXA-66 class D β-lactamases in Acinetobacter baumannii isolates from companion animals. Int J Antimicrob Agents 2017;49:37−44.ArticlePubMed
- 56. Herivaux A, Pailhories H, Quinqueneau C, et al. First report of carbapenemase-producing Acinetobacter baumannii carriage in pets from the community in France. Int J Antimicrob Agents 2016;48:220−1.ArticlePubMed
- 57. Ewers C, Klotz P, Scheufen S, et al. Genome sequence of OXA-23 producing Acinetobacter baumannii IHIT7853, a carbapenem-resistant strain from a cat belonging to international clone IC1. Gut Pathog 2016;8:37. ArticlePubMedPMC
- 58. Pomba C, Endimiani A, Rossano A, et al. First report of OXA-23-mediated carbapenem resistance in sequence type 2 multidrug-resistant Acinetobacter baumannii associated with urinary tract infection in a cat. Antimicrob Agents Chemother 2014;58:1267−8.ArticlePubMedPMCPDF
- 59. Gentilini F, Turba ME, Pasquali F, et al. Hospitalized pets as a source of carbapenem-resistance. Front Microbiol 2018;9:2872. ArticlePubMedPMC
- 60. Tyson GH, Li C, Ceric O, et al. Complete genome sequence of a carbapenem-resistant Escherichia coli isolate with blaNDM-5 from a dog in the United States. Microbiol Resour Announc 2019;8:e00872−19.ArticlePubMedPMCPDF
- 61. Hyun JE, Chung TH, Hwang CY. Identification of VIM-2 metallo-β-lactamase-producing Pseudomonas aeruginosa isolated from dogs with pyoderma and otitis in Korea. Vet Dermatol 2018;29:186. ArticlePubMedPDF
- 62. Misic D, Asanin J, Spergser J, et al. OXA-72-mediated carbapenem resistance in sequence type 1 multidrug (colistin)-resistant Acinetobacter baumannii associated with urinary tract infection in a dog from Serbia. Antimicrob Agents Chemother 2018;62:e00219−18.ArticlePubMedPMCPDF
- 63. Klotz P, Jacobmeyer L, Leidner U, et al. Acinetobacter pittii from companion animals coharboring blaOXA-58, the tet(39) region, and other resistance genes on a single plasmid. Antimicrob Agents Chemother 2017;62:e01993−17.ArticlePubMedPMCPDF
- 64. Lupo A, Chatre P, Ponsin C, et al. Clonal spread of Acinetobacter baumannii sequence type 25 carrying blaOXA-23 in companion animals in France. Antimicrob Agents Chemother 2016;61:e01881−16.ArticlePubMedPMCPDF
- 65. Kimura Y, Miyamoto T, Aoki K, et al. Analysis of IMP-1 type metallo-β-lactamase-producing Acinetobacter radioresistens isolated from companion animals. J Infect Chemother 2017;23:655−7.ArticlePubMed
- 66. Taj Z, Rasool MH, Almatroudi A, et al. Extensively drug-resistant Acinetobacter baumannii belonging to international clone II from a pet cat with urinary tract infection; the first report from Pakistan. Pol J Microbiol 2020;69:1−4.Article
- 67. Bandyopadhyay S, Banerjee J, Bhattacharyya D, et al. Companion animals emerged as an important reservoir of carbapenem-resistant Enterobacteriaceae: a report from India. Curr Microbiol 2021;78:1006−16.ArticlePubMedPDF
- 68. Oh JY, Sum S, Song WK, et al. Emergence of blaNDM-5-producing Escherichia coli ST410 in companion dogs treated with meropenem. Pak Vet J 2020;40:534−6.Article
- 69. Cole SD, Peak L, Tyson GH, et al. New Delhi Metallo-β-lactamase-5-producing Escherichia coli in companion animals, United States. Emerg Infect Dis 2020;26:381−3.ArticlePubMedPMC
- 70. Ceric O, Tyson GH, Goodman LB, et al. Enhancing the one health initiative by using whole genome sequencing to monitor antimicrobial resistance of animal pathogens: Vet-LIRN collaborative project with veterinary diagnostic laboratories in United States and Canada. BMC Vet Res 2019;15:130. ArticlePubMedPMCPDF
- 71. United States Department of Agriculture, Animal Health and Plant Inspection Services. USDA APHIS VS National Animal Health Laboratory Network (NAHLN) antimicrobial resistance pilot project. Year 1 report: 2018 [Internet]. United States Department of Agriculture: Animal Health and Plant Inspection Services; 2018 [cited 2022 Jun 22]. Available from: https://www.aphis.usda.gov/animal_health/nahln/downloads/2018%20APHIS%20AMR%20Pilot%20Project%20EOY%20Report-05.01.2019.pdf.
- 72. Marco-Fuertes A, Marin C, Lorenzo-Rebenaque L, et al. Antimicrobial resistance in companion animals: a new challenge for the One Health approach in the European Union. Vet Sci 2022;9:208. ArticlePubMedPMC
- 73. Mader R; EU-JAMRAI, Bourely C, et al. Defining the scope of the European Antimicrobial Resistance Surveillance network in Veterinary medicine (EARS-Vet): a bottom-up and One Health approach. J Antimicrob Chemother 2022;77:816−26.PubMedPMC
- 74. Guardabassi L, Schwarz S, Lloyd DH. Pet animals as reservoirs of antimicrobial-resistant bacteria. J Antimicrob Chemother 2004;54:321−32.PubMed
- 75. Committee for Medicinal Products for Veterinary Use (CVMP). Reflection paper on the risk of antimicrobial resistance transfer from companion animals [Internet]. London: European Medicines Agency; 2015 [cited 2021 Jul 20]. Available from: https://www.ema.europa.eu/en/documents/scientific-guideline/reflection-paper-risk-antimicrobial-resistance-transfer-companion-animals_en.pdf.
- 76. Bandyopadhyay S, Samanta I. Antimicrobial resistance in agri-food chain and companion animals as a re-emerging menace in post-COVID epoch: low-and middle-income countries perspective and mitigation strategies. Front Vet Sci 2020;7:620. ArticlePubMedPMC
- 77. Maseda E, Salgado P, Anillo V, et al. Risk factors for colonization by carbapenemase-producing enterobacteria at admission to a Surgical ICU: a retrospective study. Enferm Infecc Microbiol Clin 2017;35:333−7.ArticlePubMed
- 78. Codjoe FS, Donkor ES. Carbapenem resistance: a review. Med Sci (Basel) 2017;6:1. ArticlePubMedPMC
- 79. Suay-Garcia B, Perez-Gracia MT. Present and future of carbapenem-resistant Enterobacteriaceae (CRE) infections. Antibiotics (Basel) 2019;8:122. ArticlePubMedPMC
- 80. Bonardi S, Pitino R. Carbapenemase-producing bacteria in food-producing animals, wildlife and environment: a challenge for human health. Ital J Food Saf 2019;8:7956. ArticlePubMedPMCPDF
- 81. Taggar G, Attiq Rheman M, Boerlin P, et al. Molecular epidemiology of carbapenemases in Enterobacteriales from humans, animals, food and the environment. Antibiotics (Basel) 2020;9:693. ArticlePubMedPMC
- 82. Nordmann P, Dortet L, Poirel L. Carbapenem resistance in Enterobacteriaceae: here is the storm! Trends Mol Med 2012;18:263−72.ArticlePubMed
- 83. Djahmi N, Dunyach-Remy C, Pantel A, et al. Epidemiology of carbapenemase-producing Enterobacteriaceae and Acinetobacter baumannii in Mediterranean countries. Biomed Res Int 2014;2014:305784. ArticlePubMedPMCPDF
- 84. Hansen GT. Continuous evolution: perspective on the epidemiology of carbapenemase resistance among Enterobacterales and other Gram-negative bacteria. Infect Dis Ther 2021;10:75−92.ArticlePubMedPMCPDF
- 85. Menchetti L, Calipari S, Mariti C, et al. Cats and dogs: best friends or deadly enemies?: what the owners of cats and dogs living in the same household think about their relationship with people and other pets. PLoS One 2020;15:e0237822.ArticlePubMedPMC
- 86. Dolejska M, Masarikova M, Dobiasova H, et al. High prevalence of Salmonella and IMP-4-producing Enterobacteriaceae in the silver gull on Five Islands, Australia. J Antimicrob Chemother 2016;71:63−70.ArticlePubMed
References
Figure & Data
References
Citations

- First report of a blaNDM-5-carrying Escherichia coli sequence type 12 isolated from a dog with pyometra in Japan
Kazuki Harada, Tadashi Miyamoto, Michiyo Sugiyama, Tetsuo Asai
Journal of Infection and Chemotherapy.2024;[Epub] CrossRef - The European Union summary report on antimicrobial resistance in zoonotic and indicator bacteria from humans, animals and food in 2021–2022
EFSA Journal.2024;[Epub] CrossRef - Epidemiological analysis and prevention strategies in response to a shigellosis cluster outbreak: a retrospective case series in an alternative school in the Republic of Korea, 2023
Yeongseo Ahn, Sunmi Jin, Gemma Park, Hye Young Lee, Hyungyong Lee, Eunkyung Shin, Junyoung Kim, Jaeil Yoo, Yuna Kim
Osong Public Health and Research Perspectives.2024; 15(1): 68. CrossRef - The European Union Summary Report on Antimicrobial Resistance in zoonotic and indicator bacteria from humans, animals and food in 2020/2021
EFSA Journal.2023;[Epub] CrossRef - Resistome-based surveillance identifies ESKAPE pathogens as the predominant gram-negative organisms circulating in veterinary hospitals
Flavia Zendri, Cajsa M. Isgren, Jane Devaney, Vanessa Schmidt, Rachel Rankin, Dorina Timofte
Frontiers in Microbiology.2023;[Epub] CrossRef - Unveiling the emergence of multidrug-resistant pathogens in exotic pets from France: a comprehensive study (2017-2019)
Sandro Cardoso, Aurélie Le Loc’h, Inês Marques, Anabela Almeida, Sérgio Sousa, Maria José Saavedra, Sofia Anastácio, Eduarda Silveira
One Health & Implementation Research.2023; 3(4): 161. CrossRef


 Cite
Cite